Tomer Cohen
ContactNet: Geometric-Based Deep Learning Model for Predicting Protein-Protein Interactions
Jun 26, 2024Abstract:Deep learning approaches achieved significant progress in predicting protein structures. These methods are often applied to protein-protein interactions (PPIs) yet require Multiple Sequence Alignment (MSA) which is unavailable for various interactions, such as antibody-antigen. Computational docking methods are capable of sampling accurate complex models, but also produce thousands of invalid configurations. The design of scoring functions for identifying accurate models is a long-standing challenge. We develop a novel attention-based Graph Neural Network (GNN), ContactNet, for classifying PPI models obtained from docking algorithms into accurate and incorrect ones. When trained on docked antigen and modeled antibody structures, ContactNet doubles the accuracy of current state-of-the-art scoring functions, achieving accurate models among its Top-10 at 43% of the test cases. When applied to unbound antibodies, its Top-10 accuracy increases to 65%. This performance is achieved without MSA and the approach is applicable to other types of interactions, such as host-pathogens or general PPIs.
Self-Supervised Dynamic Networks for Covariate Shift Robustness
Jun 06, 2020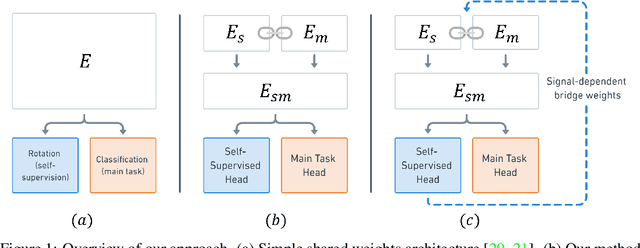

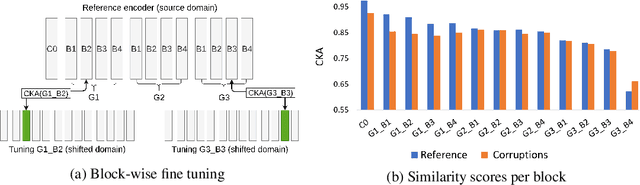
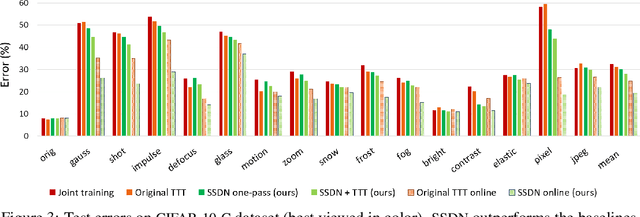
Abstract:As supervised learning still dominates most AI applications, test-time performance is often unexpected. Specifically, a shift of the input covariates, caused by typical nuisances like background-noise, illumination variations or transcription errors, can lead to a significant decrease in prediction accuracy. Recently, it was shown that incorporating self-supervision can significantly improve covariate shift robustness. In this work, we propose Self-Supervised Dynamic Networks (SSDN): an input-dependent mechanism, inspired by dynamic networks, that allows a self-supervised network to predict the weights of the main network, and thus directly handle covariate shifts at test-time. We present the conceptual and empirical advantages of the proposed method on the problem of image classification under different covariate shifts, and show that it significantly outperforms comparable methods.
A combination of 'pooling' with a prediction model can reduce by 73% the number of COVID-19 tests
May 12, 2020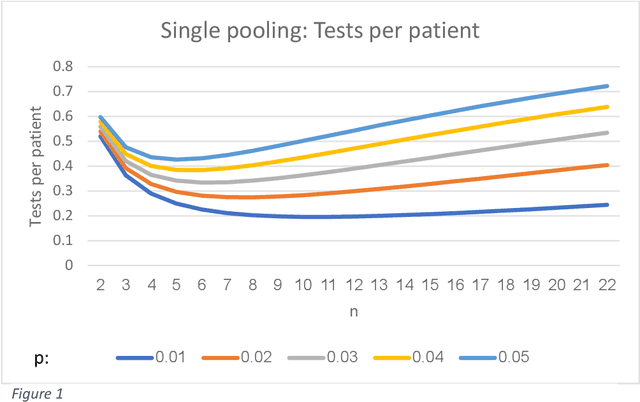
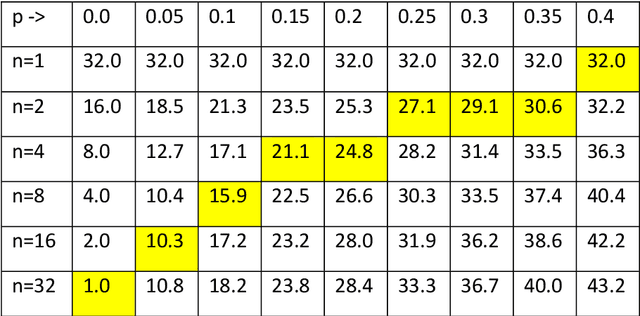
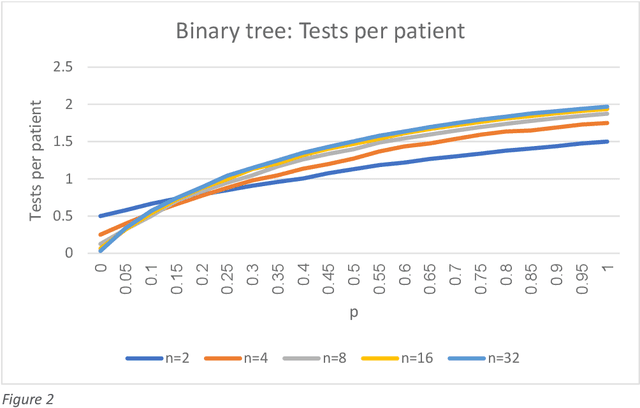
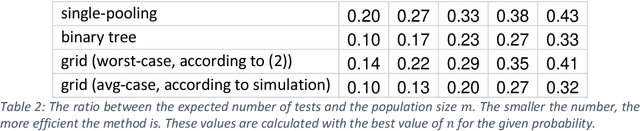
Abstract:We show that combining a prediction model (based on neural networks), with a new method of test pooling (better than the original Dorfman method, and better than double-pooling) called 'Grid', we can reduce the number of Covid-19 tests by 73%.
Bidirectional One-Shot Unsupervised Domain Mapping
Sep 04, 2019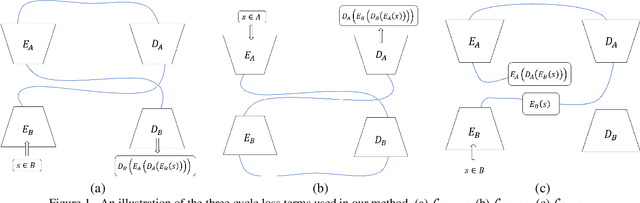
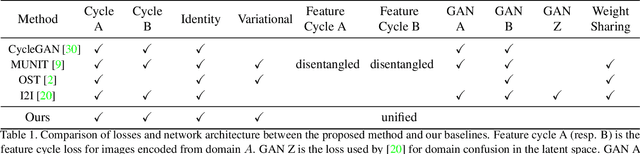
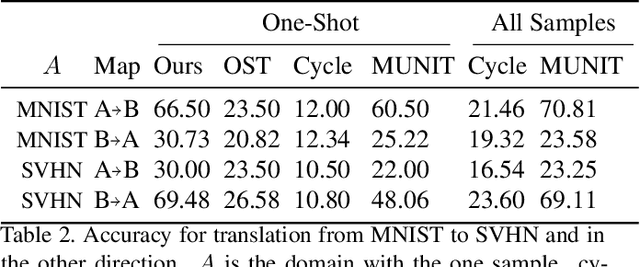

Abstract:We study the problem of mapping between a domain $A$, in which there is a single training sample and a domain $B$, for which we have a richer training set. The method we present is able to perform this mapping in both directions. For example, we can transfer all MNIST images to the visual domain captured by a single SVHN image and transform the SVHN image to the domain of the MNIST images. Our method is based on employing one encoder and one decoder for each domain, without utilizing weight sharing. The autoencoder of the single sample domain is trained to match both this sample and the latent space of domain $B$. Our results demonstrate convincing mapping between domains, where either the source or the target domain are defined by a single sample, far surpassing existing solutions. Our code is made publicly available at https://github.com/tomercohen11/BiOST
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge