Timothy J. Hall
Incorporating Gradient Similarity for Robust Time Delay Estimation in Ultrasound Elastography
Mar 30, 2022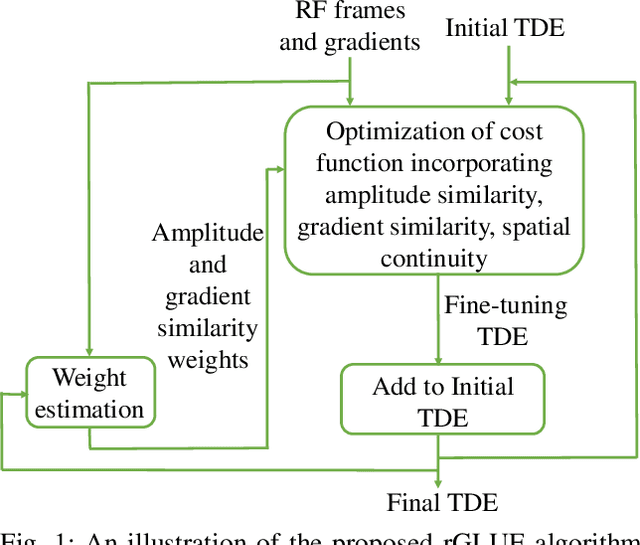
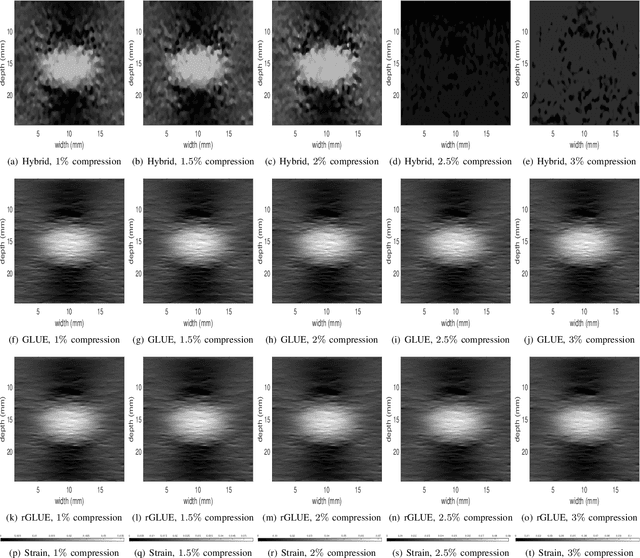
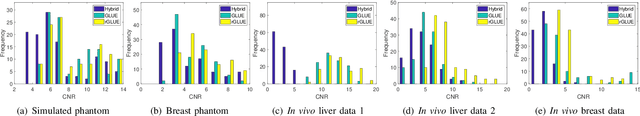
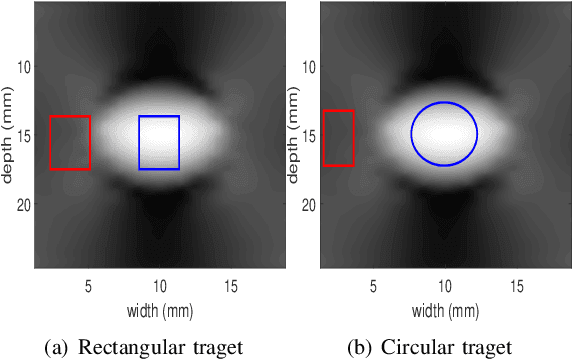
Abstract:Energy-based ultrasound elastography techniques minimize a regularized cost function consisting of data and continuity terms to obtain local displacement estimates based on the local time-delay estimation (TDE) between radio-frequency (RF) frames. The data term associated with the existing techniques takes only the amplitude similarity into account and hence is not sufficiently robust to the outlier samples present in the RF frames under consideration. This drawback creates noticeable artifacts in the strain image. To resolve this issue, we propose to formulate the data function as a linear combination of the amplitude and gradient similarity constraints. We estimate the adaptive weight concerning each similarity term following an iterative scheme. Finally, we optimize the non-linear cost function in an efficient manner to convert the problem to a sparse system of linear equations which are solved for millions of variables. We call our technique rGLUE: robust data term in GLobal Ultrasound Elastography. rGLUE has been validated using simulation, phantom, in vivo liver, and breast datasets. In all of our experiments, rGLUE substantially outperforms the recent elastography methods both visually and quantitatively. For simulated, phantom, and in vivo datasets, respectively, rGLUE achieves 107%, 18%, and 23% improvements of signal-to-noise ratio (SNR) and 61%, 19%, and 25% improvements of contrast-to-noise ratio (CNR) over GLUE, a recently-published elastography algorithm.
SweiNet: Deep Learning Based Uncertainty Quantification for Ultrasound Shear Wave Elasticity Imaging
Mar 21, 2022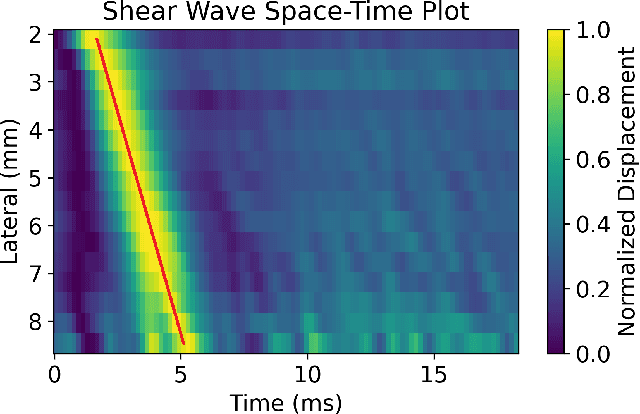
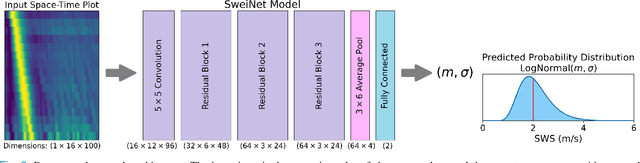
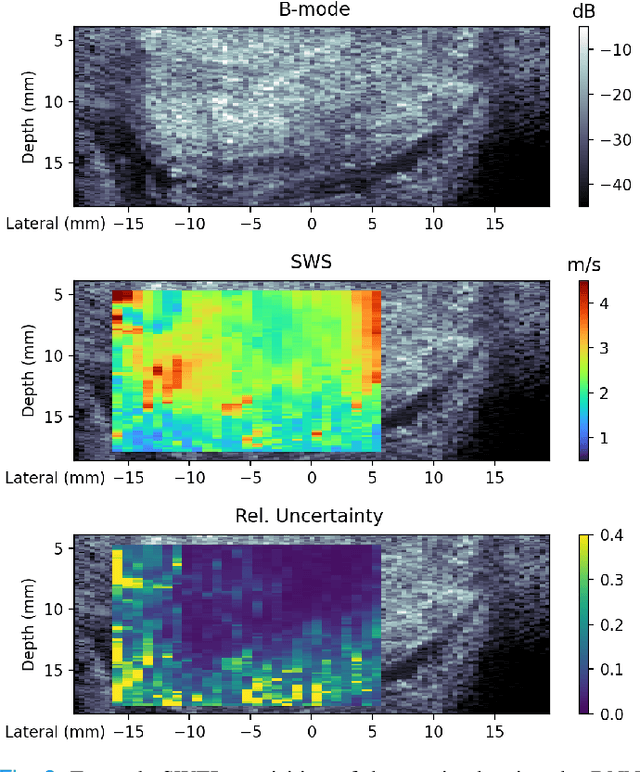
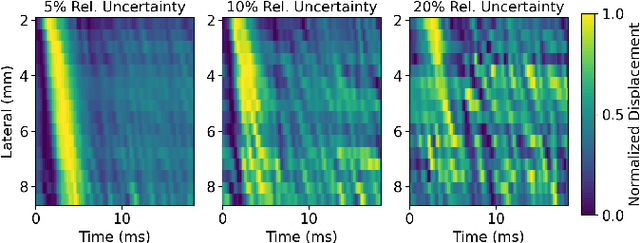
Abstract:In ultrasound shear wave elasticity (SWE) imaging, a number of algorithms exist for estimating the shear wave speed (SWS) from spatiotemporal displacement data. However, no method provides a well-calibrated and practical uncertainty metric, hindering SWE's clinical adoption and utility in downstream decision-making. Here, we designed a deep learning SWS estimator that simultaneously outputs a quantitative and well-calibrated uncertainty value for each estimate. Our deep neural network (DNN) takes as input a single 2D spatiotemporal plane of tracked displacement data and outputs the two parameters $m$ and $\sigma$ of a log-normal probability distribution. For training and testing, we used in vivo 2D-SWE data of the cervix collected from 30 pregnant subjects, totaling 551 acquisitions and >2 million space-time plots. Points were grouped by uncertainty into bins to assess uncertainty calibration: the predicted uncertainty closely matched the root-mean-square estimation error, with an average absolute percent deviation of 3.84%. We created a leave-one-out ensemble model that estimated uncertainty with better calibration (1.45%) than any individual ensemble member on a held-out patient's data. Lastly, we applied the DNN to an external dataset to evaluate its generalizability. We have made the trained model, SweiNet, openly available to provide the research community with a fast SWS estimator that also outputs a well-calibrated estimate of the predictive uncertainty.
Estimation of the Scatterer Size Distributions in Quantitative Ultrasound Using Constrained Optimization
Sep 21, 2021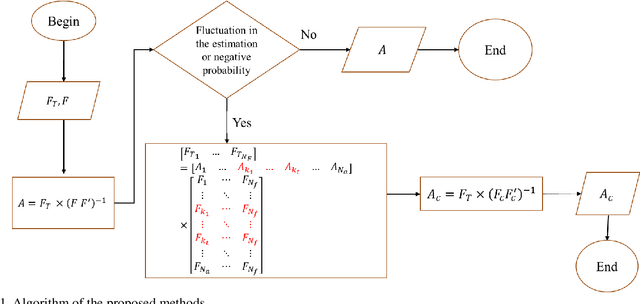
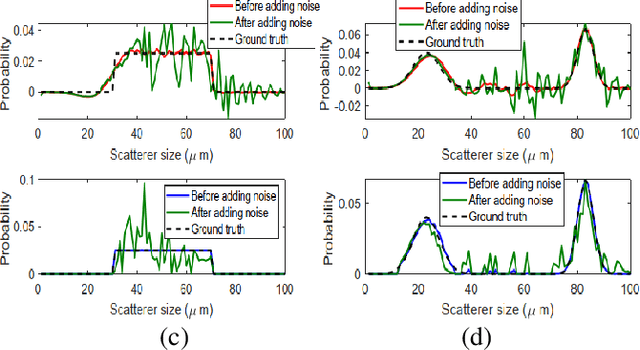
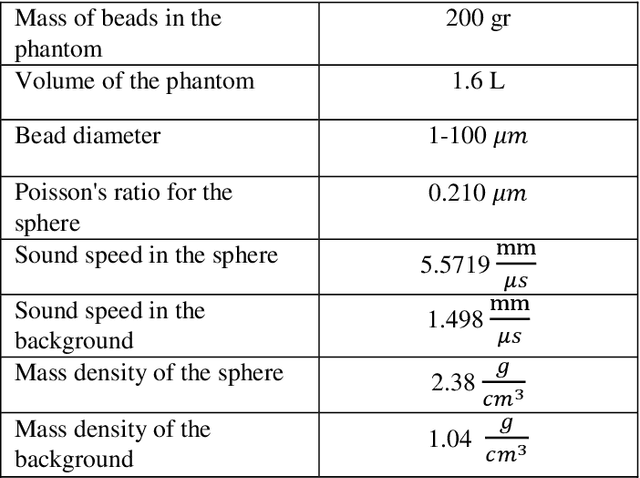
Abstract:Quantitative ultrasound (QUS) parameters such as the effective scatterer diameter (ESD) reveal tissue properties by analyzing ultrasound backscattered echo signal. ESD can be attained through parametrizing backscatter coefficient using form factor models. However, reporting a single scatterer size cannot accurately characterize a tissue, particularly when the media contains scattering sources with a broad range of sizes. Here we estimate the probability of contribution of each scatterer size by modeling the measured form factor as a linear combination of form factors from individual sacatterer sizes. We perform the estimation using two novel techniques. In the first technique, we cast scatterer size distribution as an optimization problem, and efficiently solve it using a linear system of equations. In the second technique, we use the solution of this system of equations to constrain the optimization function, and solve the constrained problem. The methods are evaluated in simulated backscattered coefficients using Faran theory. We evaluate the robustness of the proposed techniques by adding Gaussian noise. The results show that both methods can accurately estimate the scatterer size distribution, and that the second method outperforms the first one.
Ultrasound Scatterer Density Classification Using Convolutional Neural Networks by Exploiting Patch Statistics
Dec 04, 2020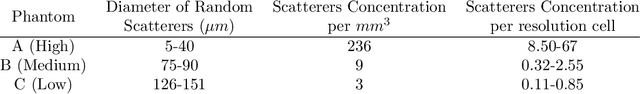
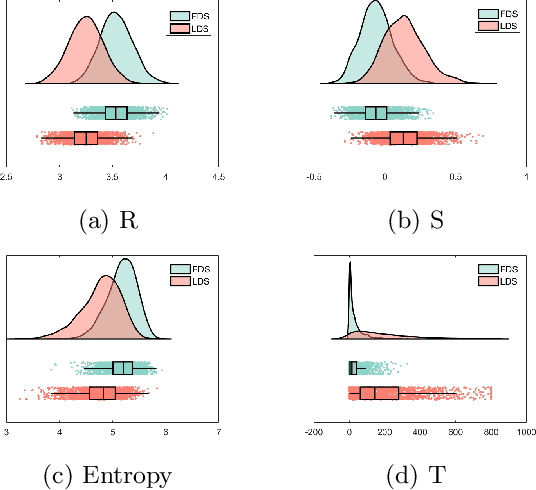
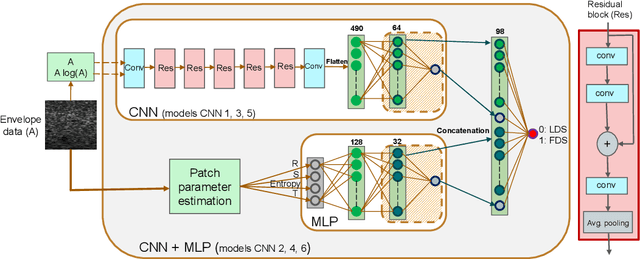
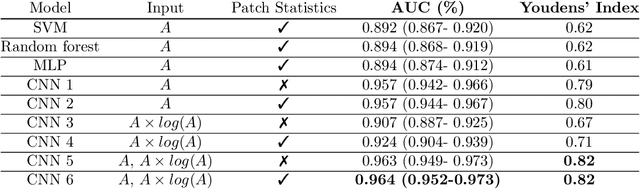
Abstract:Quantitative ultrasound (QUS) can reveal crucial information on tissue properties such as scatterer density. If the scatterer density per resolution cell is above or below 10, the tissue is considered as fully developed speckle (FDS) or low-density scatterers (LDS), respectively. Conventionally, the scatterer density has been classified using estimated statistical parameters of the amplitude of backscattered echoes. However, if the patch size is small, the estimation is not accurate. These parameters are also highly dependent on imaging settings. In this paper, we propose a convolutional neural network (CNN) architecture for QUS, and train it using simulation data. We further improve the network performance by utilizing patch statistics as additional input channels. We evaluate the network using simulation data, experimental phantoms and in vivo data. We also compare our proposed network with different classic and deep learning models, and demonstrate its superior performance in classification of tissues with different scatterer density values. The results also show that the proposed network is able to work with different imaging parameters with no need for a reference phantom. This work demonstrates the potential of CNNs in classifying scatterer density in ultrasound images.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge