Mark L. Palmeri
SweiNet: Deep Learning Based Uncertainty Quantification for Ultrasound Shear Wave Elasticity Imaging
Mar 21, 2022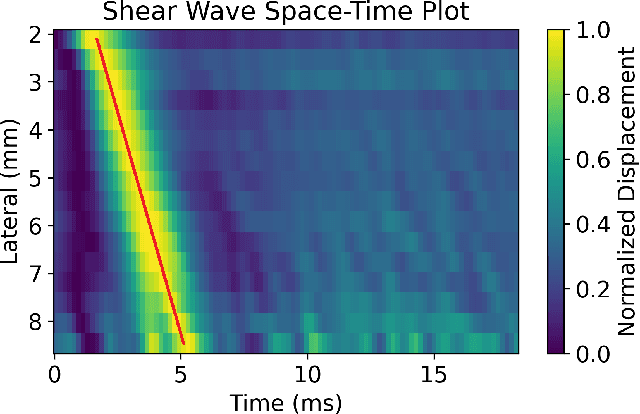
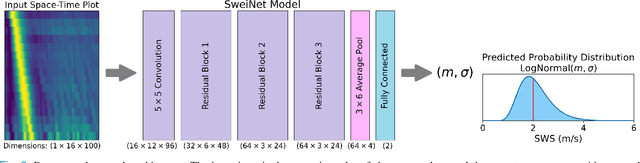
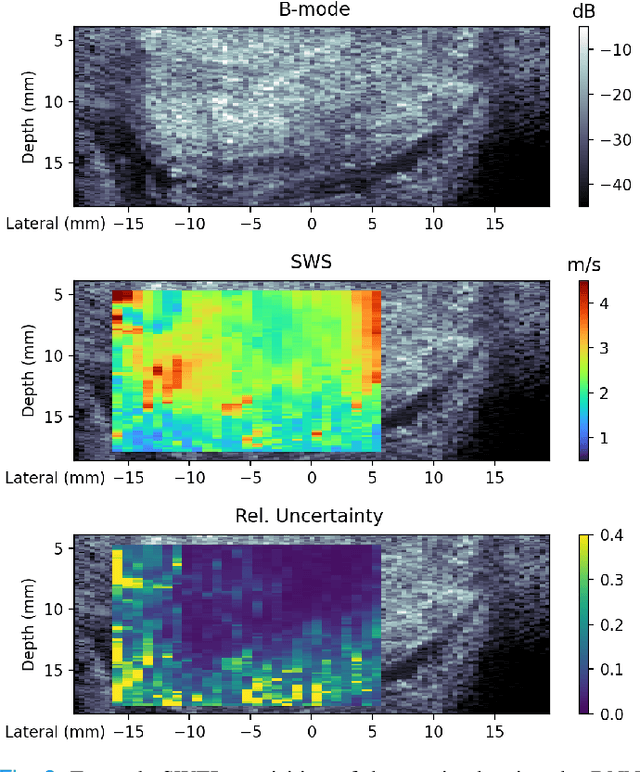
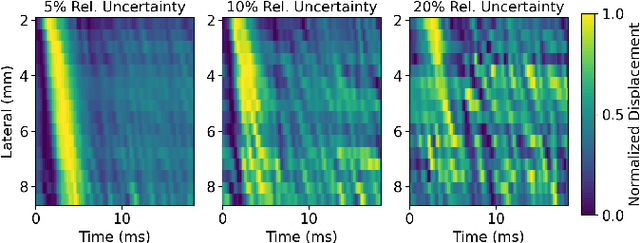
Abstract:In ultrasound shear wave elasticity (SWE) imaging, a number of algorithms exist for estimating the shear wave speed (SWS) from spatiotemporal displacement data. However, no method provides a well-calibrated and practical uncertainty metric, hindering SWE's clinical adoption and utility in downstream decision-making. Here, we designed a deep learning SWS estimator that simultaneously outputs a quantitative and well-calibrated uncertainty value for each estimate. Our deep neural network (DNN) takes as input a single 2D spatiotemporal plane of tracked displacement data and outputs the two parameters $m$ and $\sigma$ of a log-normal probability distribution. For training and testing, we used in vivo 2D-SWE data of the cervix collected from 30 pregnant subjects, totaling 551 acquisitions and >2 million space-time plots. Points were grouped by uncertainty into bins to assess uncertainty calibration: the predicted uncertainty closely matched the root-mean-square estimation error, with an average absolute percent deviation of 3.84%. We created a leave-one-out ensemble model that estimated uncertainty with better calibration (1.45%) than any individual ensemble member on a held-out patient's data. Lastly, we applied the DNN to an external dataset to evaluate its generalizability. We have made the trained model, SweiNet, openly available to provide the research community with a fast SWS estimator that also outputs a well-calibrated estimate of the predictive uncertainty.
MimickNet, Matching Clinical Post-Processing Under Realistic Black-Box Constraints
Aug 15, 2019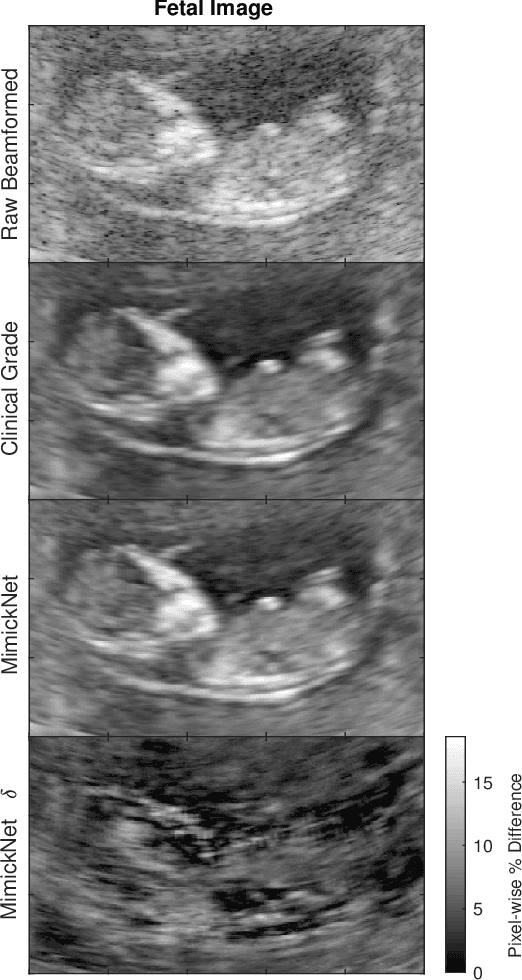
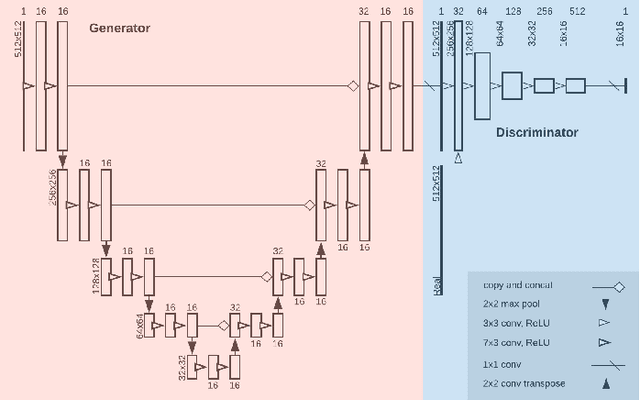
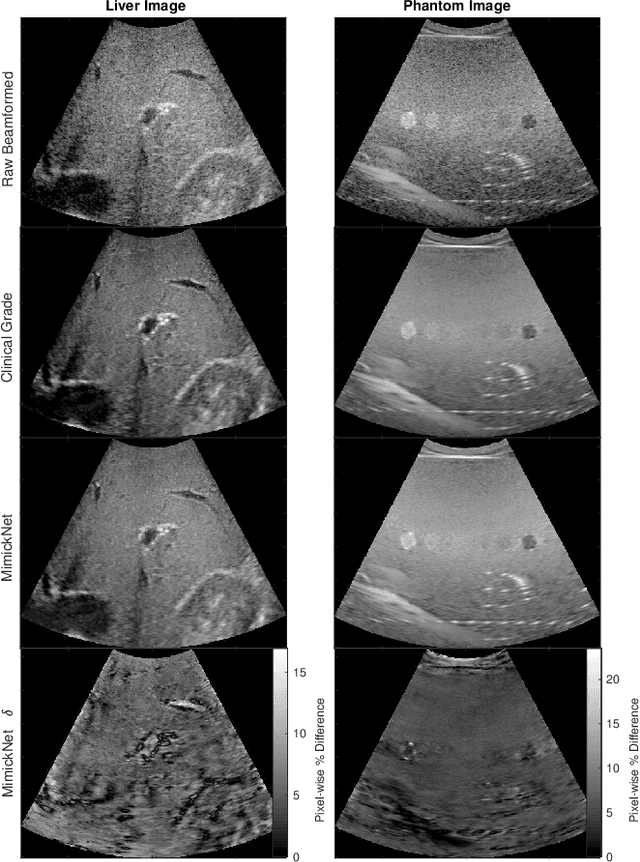
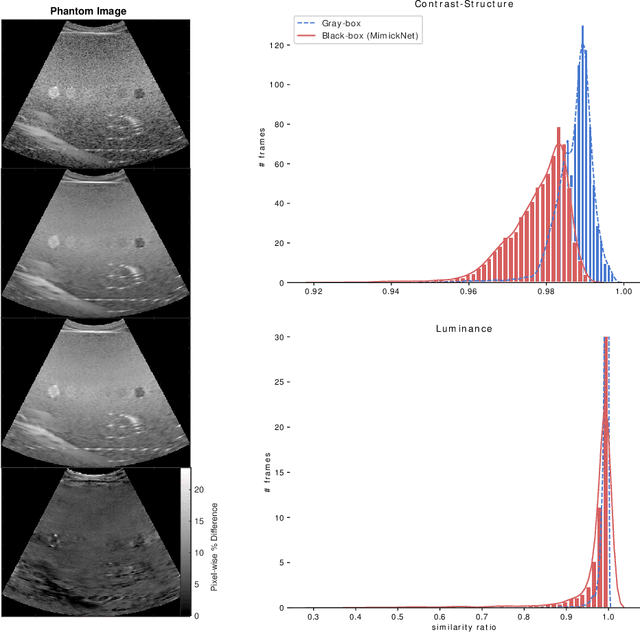
Abstract:Image post-processing is used in clinical-grade ultrasound scanners to improve image quality (e.g., reduce speckle noise and enhance contrast). These post-processing techniques vary across manufacturers and are generally kept proprietary, which presents a challenge for researchers looking to match current clinical-grade workflows. We introduce a deep learning framework, MimickNet, that transforms raw conventional delay-and-summed (DAS) beams into the approximate post-processed images found on clinical-grade scanners. Training MimickNet only requires post-processed image samples from a scanner of interest without the need for explicit pairing to raw DAS data. This flexibility allows it to hypothetically approximate any manufacturer's post-processing without access to the pre-processed data. MimickNet generates images with an average similarity index measurement (SSIM) of 0.930$\pm$0.0892 on a 300 cineloop test set, and it generalizes to cardiac cineloops outside of our train-test distribution achieving an SSIM of 0.967$\pm$0.002. We also explore the theoretical SSIM achievable by evaluating MimickNet performance when trained under gray-box constraints (i.e., when both pre-processed and post-processed images are available). To our knowledge, this is the first work to establish deep learning models that closely approximate current clinical-grade ultrasound post-processing under realistic black-box constraints where before and after post-processing data is unavailable. MimickNet serves as a clinical post-processing baseline for future works in ultrasound image formation to compare against. To this end, we have made the MimickNet software open source.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge