Tilendra Choudhary
Deep Representation Learning-Based Dynamic Trajectory Phenotyping for Acute Respiratory Failure in Medical Intensive Care Units
May 04, 2024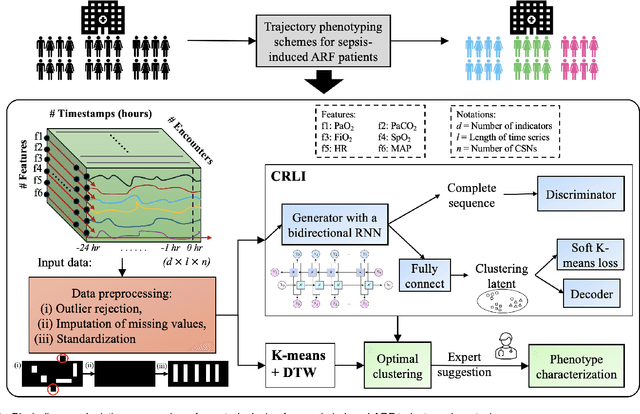
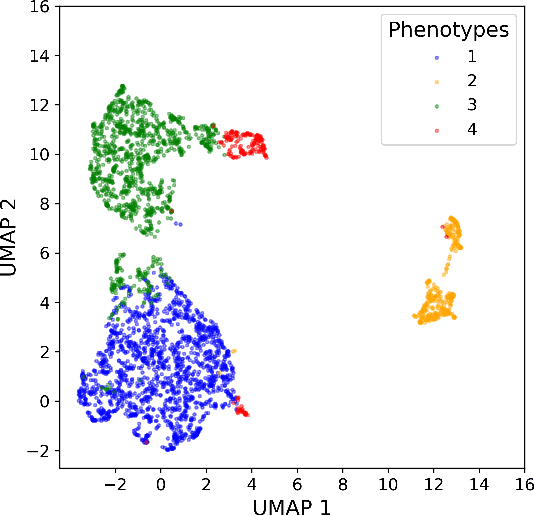
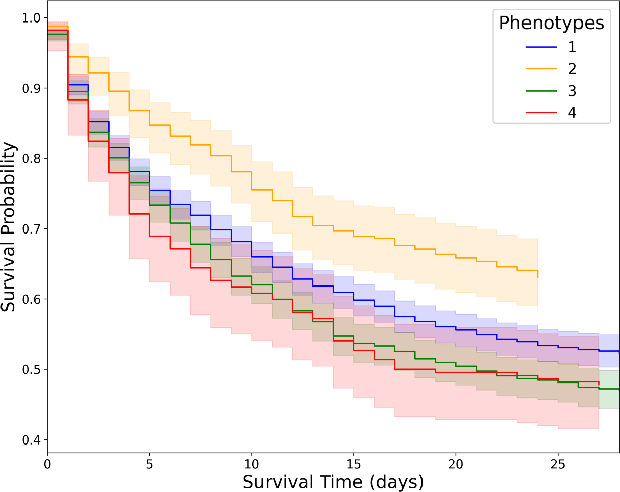
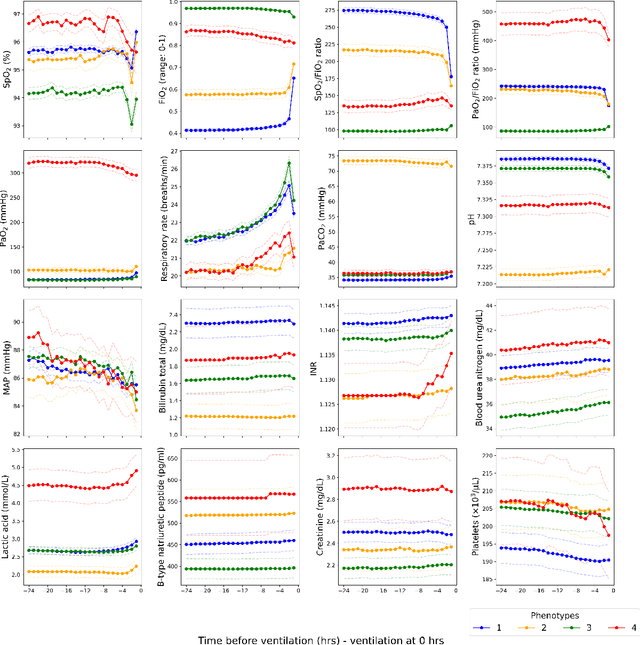
Abstract:Sepsis-induced acute respiratory failure (ARF) is a serious complication with a poor prognosis. This paper presents a deep representation learningbased phenotyping method to identify distinct groups of clinical trajectories of septic patients with ARF. For this retrospective study, we created a dataset from electronic medical records (EMR) consisting of data from sepsis patients admitted to medical intensive care units who required at least 24 hours of invasive mechanical ventilation at a quarternary care academic hospital in southeast USA for the years 2016-2021. A total of N=3349 patient encounters were included in this study. Clustering Representation Learning on Incomplete Time Series Data (CRLI) algorithm was applied to a parsimonious set of EMR variables in this data set. To validate the optimal number of clusters, the K-means algorithm was used in conjunction with dynamic time warping. Our model yielded four distinct patient phenotypes that were characterized as liver dysfunction/heterogeneous, hypercapnia, hypoxemia, and multiple organ dysfunction syndrome by a critical care expert. A Kaplan-Meier analysis to compare the 28-day mortality trends exhibited significant differences (p < 0.005) between the four phenotypes. The study demonstrates the utility of our deep representation learning-based approach in unraveling phenotypes that reflect the heterogeneity in sepsis-induced ARF in terms of different mortality outcomes and severity. These phenotypes might reveal important clinical insights into an effective prognosis and tailored treatment strategies.
Robust Meta-Model for Predicting the Need for Blood Transfusion in Non-traumatic ICU Patients
Jan 01, 2024Abstract:Objective: Blood transfusions, crucial in managing anemia and coagulopathy in ICU settings, require accurate prediction for effective resource allocation and patient risk assessment. However, existing clinical decision support systems have primarily targeted a particular patient demographic with unique medical conditions and focused on a single type of blood transfusion. This study aims to develop an advanced machine learning-based model to predict the probability of transfusion necessity over the next 24 hours for a diverse range of non-traumatic ICU patients. Methods: We conducted a retrospective cohort study on 72,072 adult non-traumatic ICU patients admitted to a high-volume US metropolitan academic hospital between 2016 and 2020. We developed a meta-learner and various machine learning models to serve as predictors, training them annually with four-year data and evaluating on the fifth, unseen year, iteratively over five years. Results: The experimental results revealed that the meta-model surpasses the other models in different development scenarios. It achieved notable performance metrics, including an Area Under the Receiver Operating Characteristic (AUROC) curve of 0.97, an accuracy rate of 0.93, and an F1-score of 0.89 in the best scenario. Conclusion: This study pioneers the use of machine learning models for predicting blood transfusion needs in a diverse cohort of critically ill patients. The findings of this evaluation confirm that our model not only predicts transfusion requirements effectively but also identifies key biomarkers for making transfusion decisions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge