Teaghan O'Briain
FlowNet-PET: Unsupervised Learning to Perform Respiratory Motion Correction in PET Imaging
May 27, 2022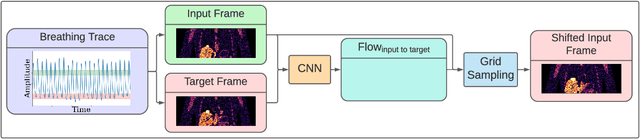
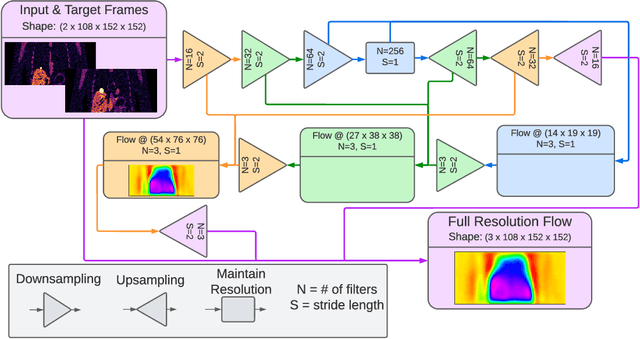
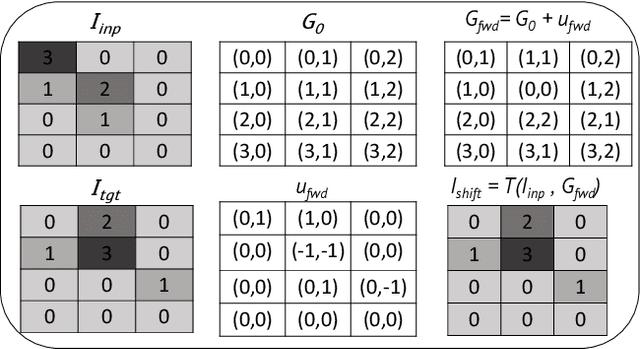
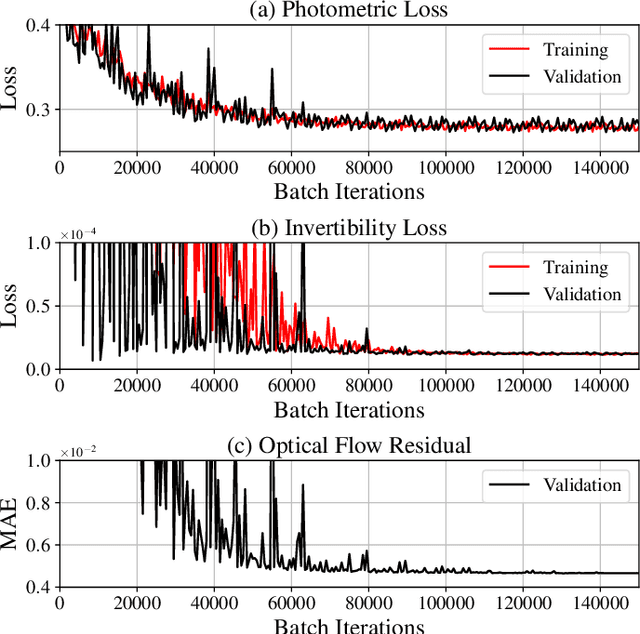
Abstract:To correct for breathing motion in PET imaging, an interpretable and unsupervised deep learning technique, FlowNet-PET, was constructed. The network was trained to predict the optical flow between two PET frames from different breathing amplitude ranges. As a result, the trained model groups different retrospectively-gated PET images together into a motion-corrected single bin, providing a final image with similar counting statistics as a non-gated image, but without the blurring effects that were initially observed. As a proof-of-concept, FlowNet-PET was applied to anthropomorphic digital phantom data, which provided the possibility to design robust metrics to quantify the corrections. When comparing the predicted optical flows to the ground truths, the median absolute error was found to be smaller than the pixel and slice widths, even for the phantom with a diaphragm movement of 21 mm. The improvements were illustrated by comparing against images without motion and computing the intersection over union (IoU) of the tumors as well as the enclosed activity and coefficient of variation (CoV) within the no-motion tumor volume before and after the corrections were applied. The average relative improvements provided by the network were 54%, 90%, and 76% for the IoU, total activity, and CoV, respectively. The results were then compared against the conventional retrospective phase binning approach. FlowNet-PET achieved similar results as retrospective binning, but only required one sixth of the scan duration. The code and data used for training and analysis has been made publicly available (https://github.com/teaghan/FlowNet_PET).
Interpreting Stellar Spectra with Unsupervised Domain Adaptation
Jul 06, 2020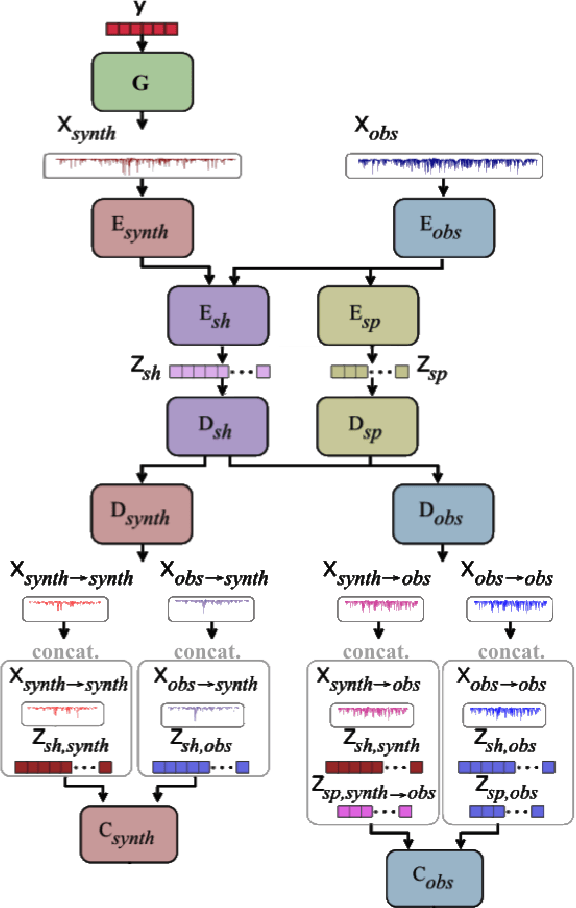
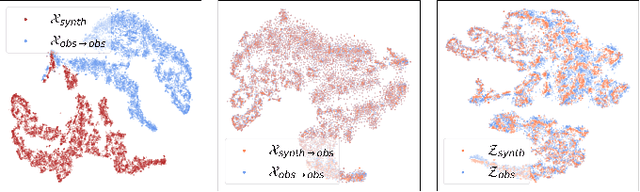
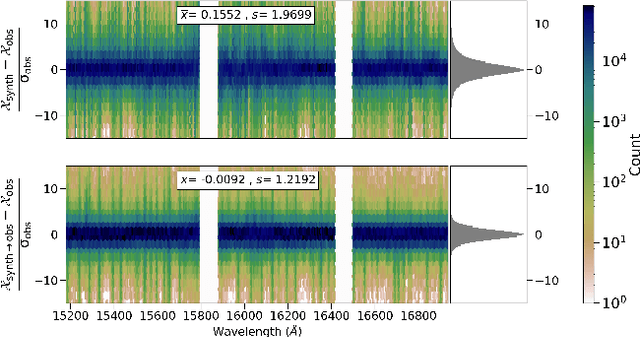
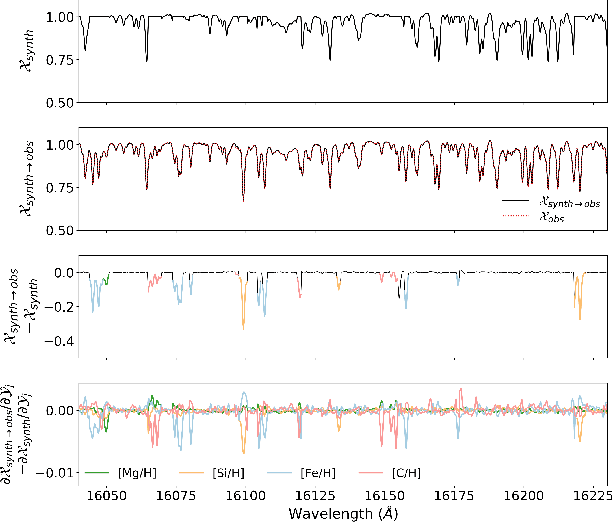
Abstract:We discuss how to achieve mapping from large sets of imperfect simulations and observational data with unsupervised domain adaptation. Under the hypothesis that simulated and observed data distributions share a common underlying representation, we show how it is possible to transfer between simulated and observed domains. Driven by an application to interpret stellar spectroscopic sky surveys, we construct the domain transfer pipeline from two adversarial autoencoders on each domains with a disentangling latent space, and a cycle-consistency constraint. We then construct a differentiable pipeline from physical stellar parameters to realistic observed spectra, aided by a supplementary generative surrogate physics emulator network. We further exemplify the potential of the method on the reconstructed spectra quality and to discover new spectral features associated to elemental abundances.
Cycle-StarNet: Bridging the gap between theory and data by leveraging large datasets
Jul 06, 2020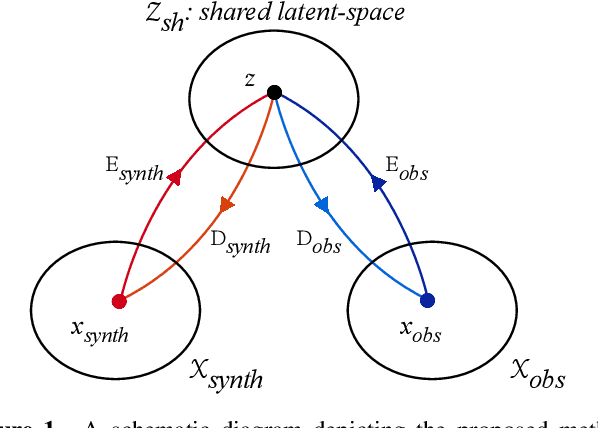
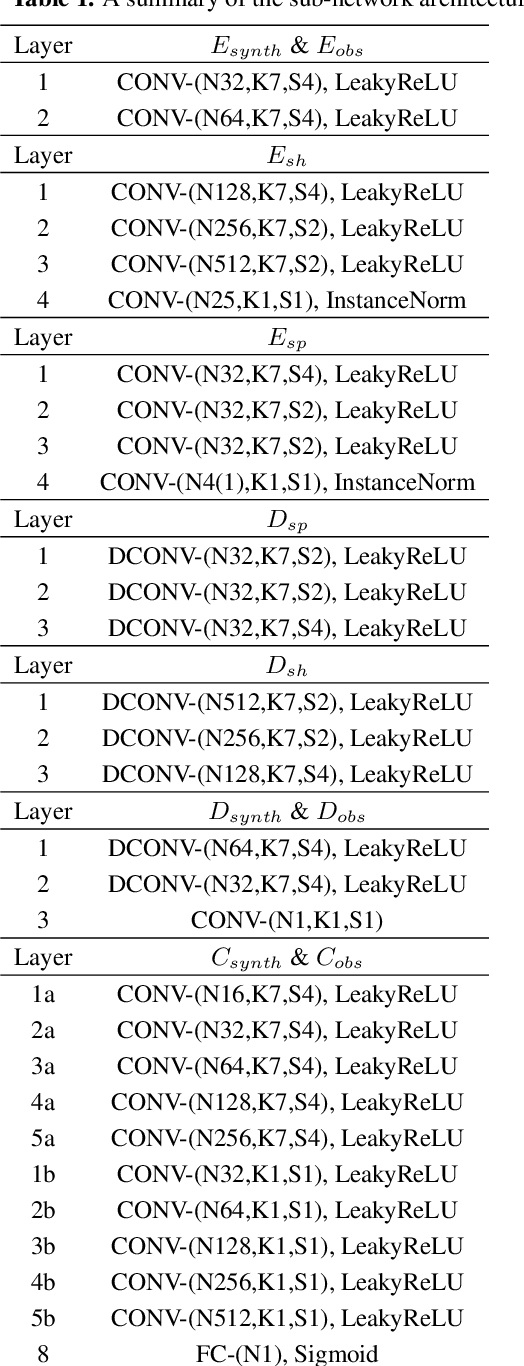
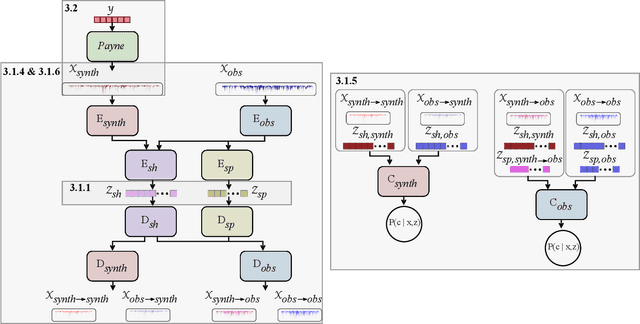
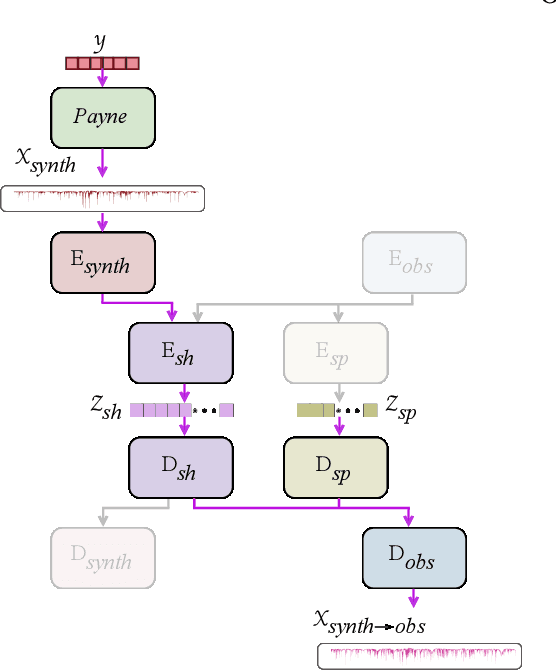
Abstract:Spectroscopy provides an immense amount of information on stellar objects, and this field continues to grow with recent developments in multi-object data acquisition and rapid data analysis techniques. Current automated methods for analyzing spectra are either (a) data-driven models, which require large amounts of data with prior knowledge of stellar parameters and elemental abundances, or (b) based on theoretical synthetic models that are susceptible to the gap between theory and practice. In this study, we present a hybrid generative domain adaptation method to turn simulated stellar spectra into realistic spectra, learning from the large spectroscopic surveys. We use a neural network to emulate computationally expensive stellar spectra simulations, and then train a separate unsupervised domain-adaptation network that learns to relate the generated synthetic spectra to observational spectra. Consequently, the network essentially produces data-driven models without the need for a labeled training set. As a proof of concept, two case studies are presented. The first of which is the auto-calibration of synthetic models without using any standard stars. To accomplish this, synthetic models are morphed into spectra that resemble observations, thereby reducing the gap between theory and observations. The second case study is the identification of the elemental source of missing spectral lines in the synthetic modelling. These sources are predicted by interpreting the differences between the domain-adapted and original spectral models. To test our ability to identify missing lines, we use a mock dataset and show that, even with noisy observations, absorption lines can be recovered when they are absent in one of the domains. While we focus on spectral analyses in this study, this method can be applied to other fields, which use large data sets and are currently limited by modelling accuracy.
Reducing the Human Effort in Developing PET-CT Registration
Nov 25, 2019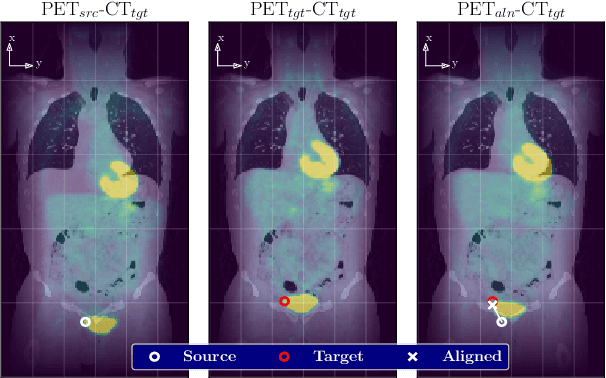

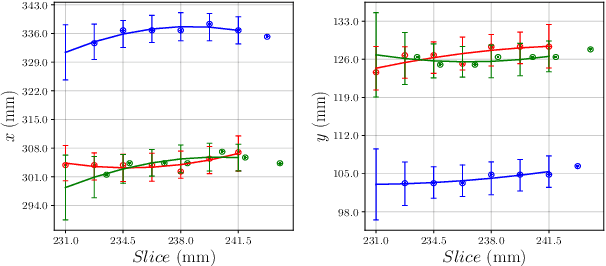
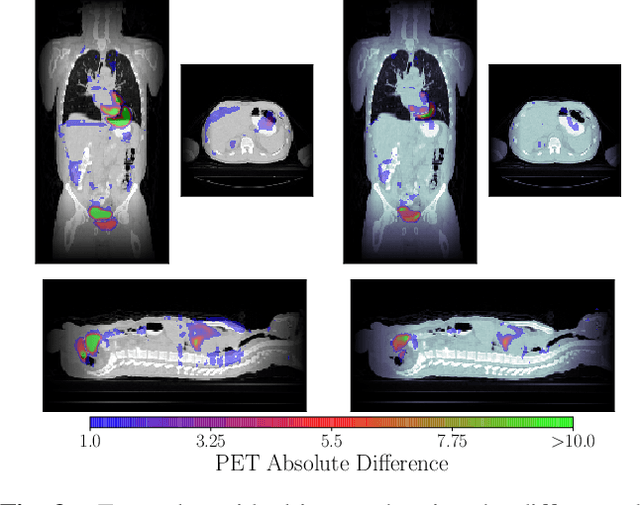
Abstract:We aim to reduce the tedious nature of developing and evaluating methods for aligning PET-CT scans from multiple patient visits. Current methods for registration rely on correspondences that are created manually by medical experts with 3D manipulation, or assisted alignments done by utilizing mutual information across CT scans that may not be consistent when transferred to the PET images. Instead, we propose to label multiple key points across several 2D slices, which we then fit a key curve to. This removes the need for creating manual alignments in 3D and makes the labelling process easier. We use these key curves to define an error metric for the alignments that can be computed efficiently. While our metric is non-differentiable, we further show that we can utilize it during the training of our deep model via a novel method. Specifically, instead of relying on detailed geometric labels -- e.g., manual 3D alignments -- we use synthetically generated deformations of real data. To incorporate robustness to changes that occur between visits other than geometric changes, we enforce consistency across visits in the deep network's internal representations. We demonstrate the potential of our method via qualitative and quantitative experiments.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge