Spiros Denaxas
Diffsurv: Differentiable sorting for censored time-to-event data
Apr 26, 2023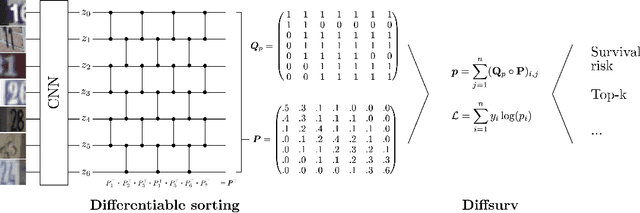

Abstract:Survival analysis is a crucial semi-supervised task in machine learning with numerous real-world applications, particularly in healthcare. Currently, the most common approach to survival analysis is based on Cox's partial likelihood, which can be interpreted as a ranking model optimized on a lower bound of the concordance index. This relation between ranking models and Cox's partial likelihood considers only pairwise comparisons. Recent work has developed differentiable sorting methods which relax this pairwise independence assumption, enabling the ranking of sets of samples. However, current differentiable sorting methods cannot account for censoring, a key factor in many real-world datasets. To address this limitation, we propose a novel method called Diffsurv. We extend differentiable sorting methods to handle censored tasks by predicting matrices of possible permutations that take into account the label uncertainty introduced by censored samples. We contrast this approach with methods derived from partial likelihood and ranking losses. Our experiments show that Diffsurv outperforms established baselines in various simulated and real-world risk prediction scenarios. Additionally, we demonstrate the benefits of the algorithmic supervision enabled by Diffsurv by presenting a novel method for top-k risk prediction that outperforms current methods.
Application of Clinical Concept Embeddings for Heart Failure Prediction in UK EHR data
Nov 28, 2018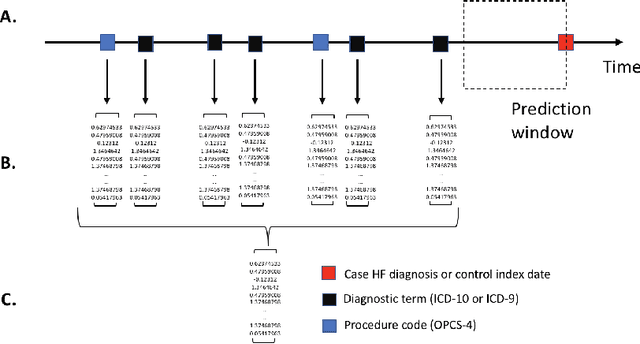
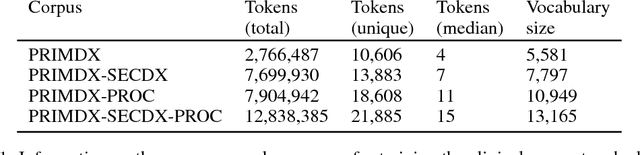
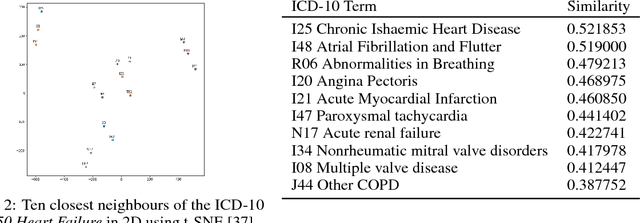
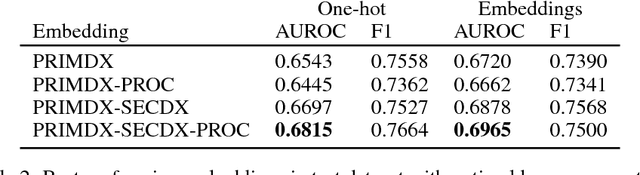
Abstract:Electronic health records (EHR) are increasingly being used for constructing disease risk prediction models. Feature engineering in EHR data however is challenging due to their highly dimensional and heterogeneous nature. Low-dimensional representations of EHR data can potentially mitigate these challenges. In this paper, we use global vectors (GloVe) to learn word embeddings for diagnoses and procedures recorded using 13 million ontology terms across 2.7 million hospitalisations in national UK EHR. We demonstrate the utility of these embeddings by evaluating their performance in identifying patients which are at higher risk of being hospitalised for congestive heart failure. Our findings indicate that embeddings can enable the creation of robust EHR-derived disease risk prediction models and address some the limitations associated with manual clinical feature engineering.
Evaluation of Semantic Web Technologies for Storing Computable Definitions of Electronic Health Records Phenotyping Algorithms
Jul 24, 2017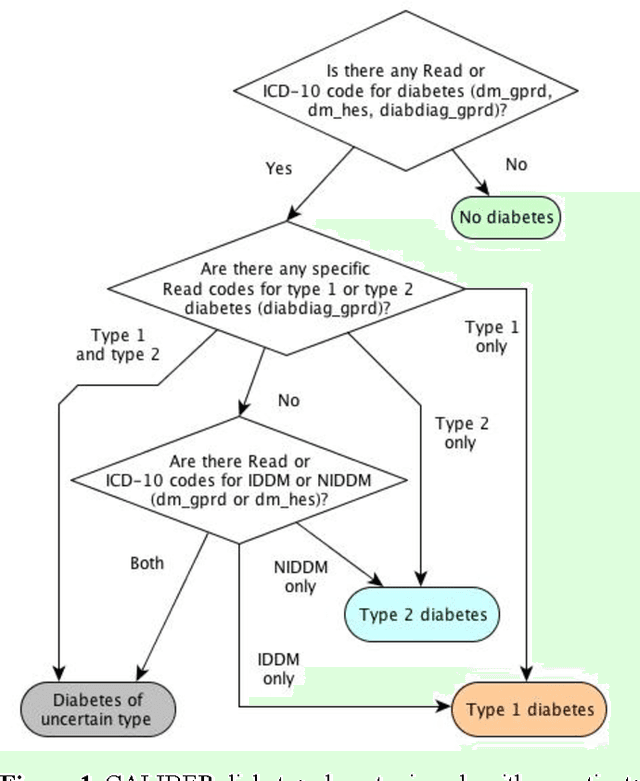
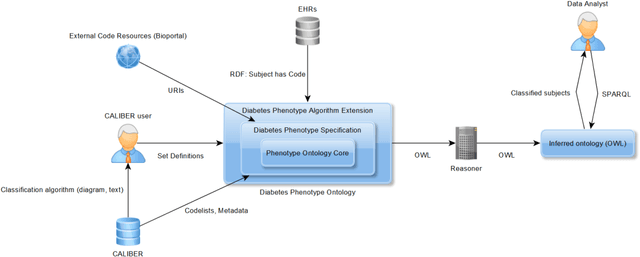
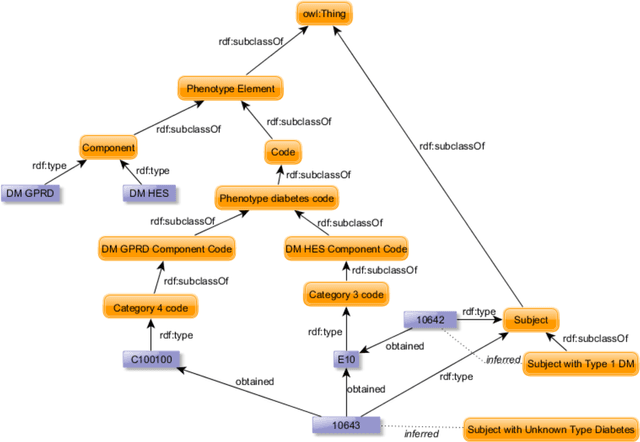
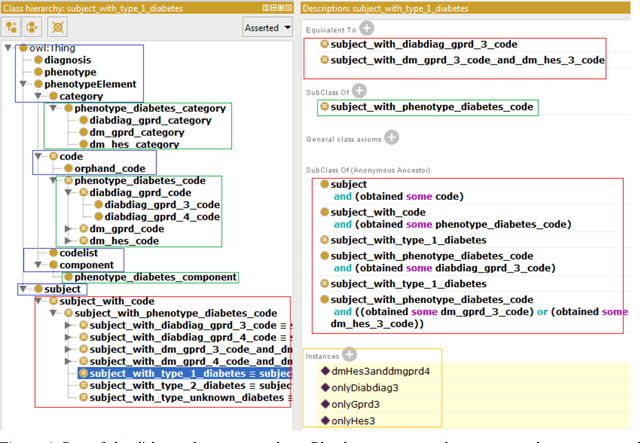
Abstract:Electronic Health Records are electronic data generated during or as a byproduct of routine patient care. Structured, semi-structured and unstructured EHR offer researchers unprecedented phenotypic breadth and depth and have the potential to accelerate the development of precision medicine approaches at scale. A main EHR use-case is defining phenotyping algorithms that identify disease status, onset and severity. Phenotyping algorithms utilize diagnoses, prescriptions, laboratory tests, symptoms and other elements in order to identify patients with or without a specific trait. No common standardized, structured, computable format exists for storing phenotyping algorithms. The majority of algorithms are stored as human-readable descriptive text documents making their translation to code challenging due to their inherent complexity and hinders their sharing and re-use across the community. In this paper, we evaluate the two key Semantic Web Technologies, the Web Ontology Language and the Resource Description Framework, for enabling computable representations of EHR-driven phenotyping algorithms.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge