Soundarya Krishnan
ReALM: Reference Resolution As Language Modeling
Mar 29, 2024
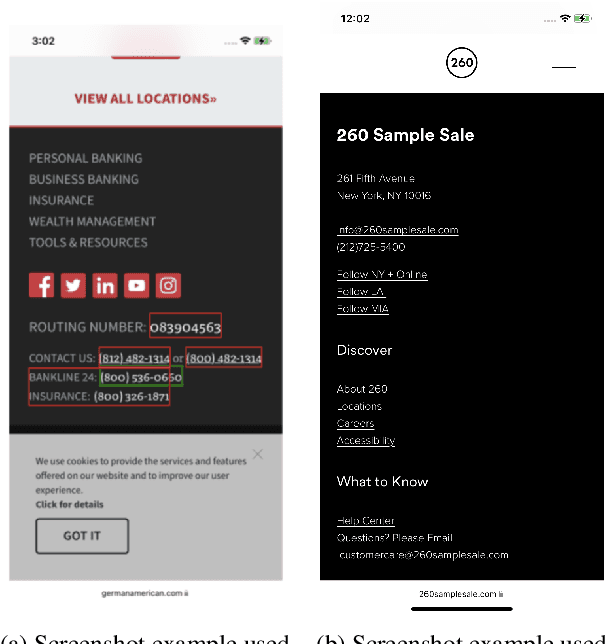
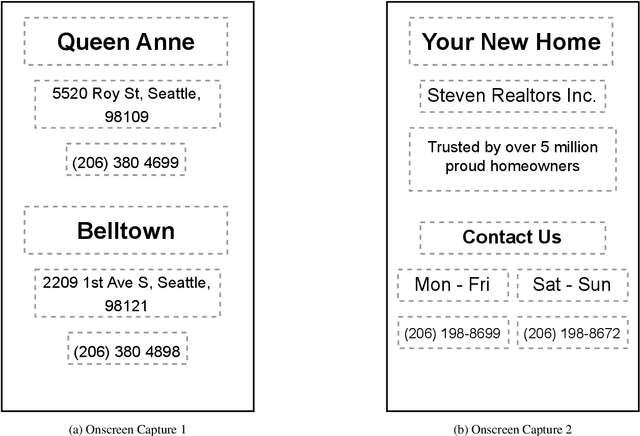
Abstract:Reference resolution is an important problem, one that is essential to understand and successfully handle context of different kinds. This context includes both previous turns and context that pertains to non-conversational entities, such as entities on the user's screen or those running in the background. While LLMs have been shown to be extremely powerful for a variety of tasks, their use in reference resolution, particularly for non-conversational entities, remains underutilized. This paper demonstrates how LLMs can be used to create an extremely effective system to resolve references of various types, by showing how reference resolution can be converted into a language modeling problem, despite involving forms of entities like those on screen that are not traditionally conducive to being reduced to a text-only modality. We demonstrate large improvements over an existing system with similar functionality across different types of references, with our smallest model obtaining absolute gains of over 5% for on-screen references. We also benchmark against GPT-3.5 and GPT-4, with our smallest model achieving performance comparable to that of GPT-4, and our larger models substantially outperforming it.
Constructing and Evaluating an Explainable Model for COVID-19 Diagnosis from Chest X-rays
Dec 19, 2020



Abstract:In this paper, our focus is on constructing models to assist a clinician in the diagnosis of COVID-19 patients in situations where it is easier and cheaper to obtain X-ray data than to obtain high-quality images like those from CT scans. Deep neural networks have repeatedly been shown to be capable of constructing highly predictive models for disease detection directly from image data. However, their use in assisting clinicians has repeatedly hit a stumbling block due to their black-box nature. Some of this difficulty can be alleviated if predictions were accompanied by explanations expressed in clinically relevant terms. In this paper, deep neural networks are used to extract domain-specific features(morphological features like ground-glass opacity and disease indications like pneumonia) directly from the image data. Predictions about these features are then used to construct a symbolic model (a decision tree) for the diagnosis of COVID-19 from chest X-rays, accompanied with two kinds of explanations: visual (saliency maps, derived from the neural stage), and textual (logical descriptions, derived from the symbolic stage). A radiologist rates the usefulness of the visual and textual explanations. Our results demonstrate that neural models can be employed usefully in identifying domain-specific features from low-level image data; that textual explanations in terms of clinically relevant features may be useful; and that visual explanations will need to be clinically meaningful to be useful.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge