Soren Brage
UDAMA: Unsupervised Domain Adaptation through Multi-discriminator Adversarial Training with Noisy Labels Improves Cardio-fitness Prediction
Jul 31, 2023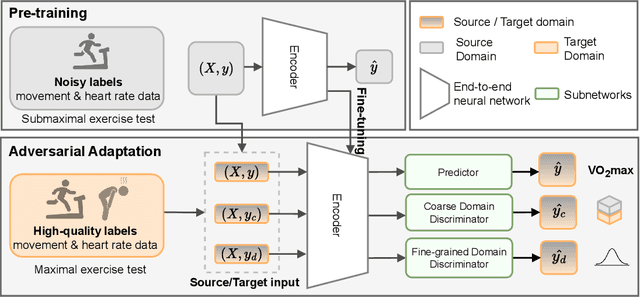
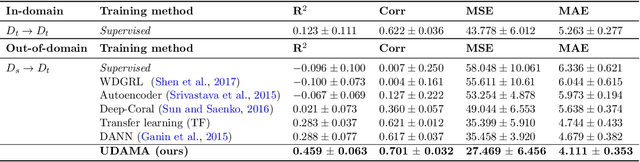
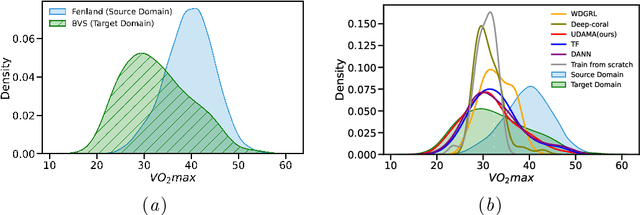

Abstract:Deep learning models have shown great promise in various healthcare monitoring applications. However, most healthcare datasets with high-quality (gold-standard) labels are small-scale, as directly collecting ground truth is often costly and time-consuming. As a result, models developed and validated on small-scale datasets often suffer from overfitting and do not generalize well to unseen scenarios. At the same time, large amounts of imprecise (silver-standard) labeled data, annotated by approximate methods with the help of modern wearables and in the absence of ground truth validation, are starting to emerge. However, due to measurement differences, this data displays significant label distribution shifts, which motivates the use of domain adaptation. To this end, we introduce UDAMA, a method with two key components: Unsupervised Domain Adaptation and Multidiscriminator Adversarial Training, where we pre-train on the silver-standard data and employ adversarial adaptation with the gold-standard data along with two domain discriminators. In particular, we showcase the practical potential of UDAMA by applying it to Cardio-respiratory fitness (CRF) prediction. CRF is a crucial determinant of metabolic disease and mortality, and it presents labels with various levels of noise (goldand silver-standard), making it challenging to establish an accurate prediction model. Our results show promising performance by alleviating distribution shifts in various label shift settings. Additionally, by using data from two free-living cohort studies (Fenland and BBVS), we show that UDAMA consistently outperforms up to 12% compared to competitive transfer learning and state-of-the-art domain adaptation models, paving the way for leveraging noisy labeled data to improve fitness estimation at scale.
Turning Silver into Gold: Domain Adaptation with Noisy Labels for Wearable Cardio-Respiratory Fitness Prediction
Nov 20, 2022Abstract:Deep learning models have shown great promise in various healthcare applications. However, most models are developed and validated on small-scale datasets, as collecting high-quality (gold-standard) labels for health applications is often costly and time-consuming. As a result, these models may suffer from overfitting and not generalize well to unseen data. At the same time, an extensive amount of data with imprecise labels (silver-standard) is starting to be generally available, as collected from inexpensive wearables like accelerometers and electrocardiography sensors. These currently underutilized datasets and labels can be leveraged to produce more accurate clinical models. In this work, we propose UDAMA, a novel model with two key components: Unsupervised Domain Adaptation and Multi-discriminator Adversarial training, which leverage noisy data from source domain (the silver-standard dataset) to improve gold-standard modeling. We validate our framework on the challenging task of predicting lab-measured maximal oxygen consumption (VO$_{2}$max), the benchmark metric of cardio-respiratory fitness, using free-living wearable sensor data from two cohort studies as inputs. Our experiments show that the proposed framework achieves the best performance of corr = 0.665 $\pm$ 0.04, paving the way for accurate fitness estimation at scale.
Longitudinal cardio-respiratory fitness prediction through free-living wearable sensors
May 06, 2022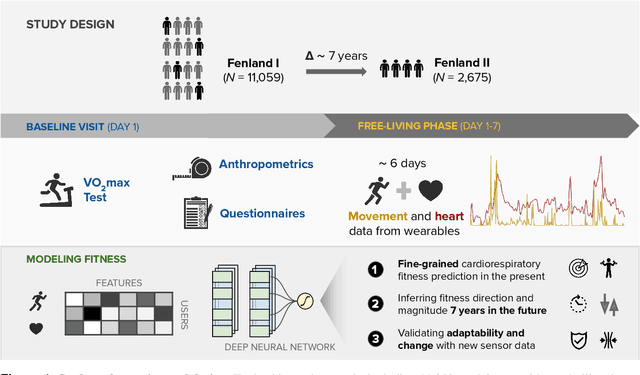
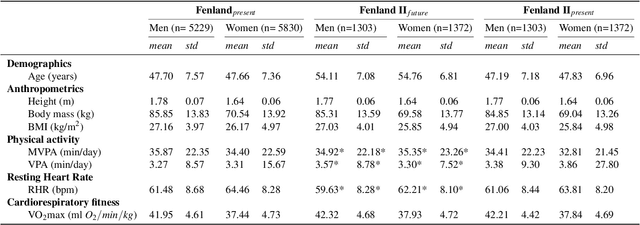
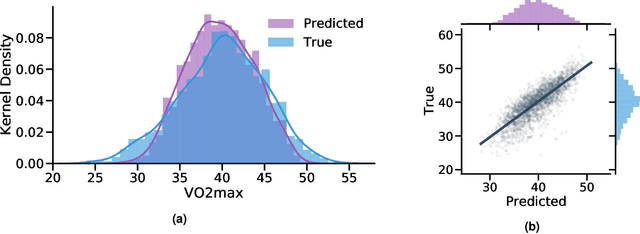
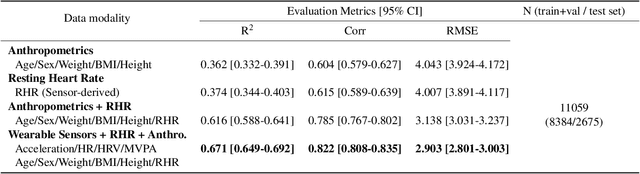
Abstract:Cardiorespiratory fitness is an established predictor of metabolic disease and mortality. Fitness is directly measured as maximal oxygen consumption (VO2max), or indirectly assessed using heart rate response to a standard exercise test. However, such testing is costly and burdensome, limiting its utility and scalability. Fitness can also be approximated using resting heart rate and self-reported exercise habits but with lower accuracy. Modern wearables capture dynamic heart rate data which, in combination with machine learning models, could improve fitness prediction. In this work, we analyze movement and heart rate signals from wearable sensors in free-living conditions from 11,059 participants who also underwent a standard exercise test, along with a longitudinal repeat cohort of 2,675 participants. We design algorithms and models that convert raw sensor data into cardio-respiratory fitness estimates, and validate these estimates' ability to capture fitness profiles in a longitudinal cohort over time while subjects engaged in real-world (non-exercise) behaviour. Additionally, we validate our methods with a third external cohort of 181 participants who underwent maximal VO2max testing, which is considered the gold standard measurement because it requires reaching one's maximum heart rate and exhaustion level. Our results show that the developed models yield a high correlation (r = 0.82, 95CI 0.80-0.83), when compared to the ground truth in a holdout sample. These models outperform conventional non-exercise fitness models and traditional bio-markers using measurements of normal daily living without the need for a specific exercise test. Additionally, we show the adaptability and applicability of this approach for detecting fitness change over time in the longitudinal subsample that repeated measurements after 7 years.
SelfHAR: Improving Human Activity Recognition through Self-training with Unlabeled Data
Feb 11, 2021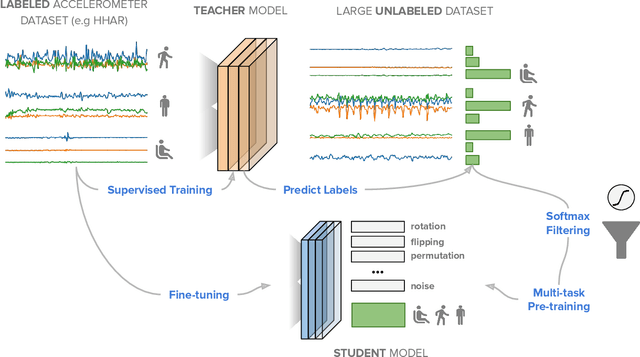
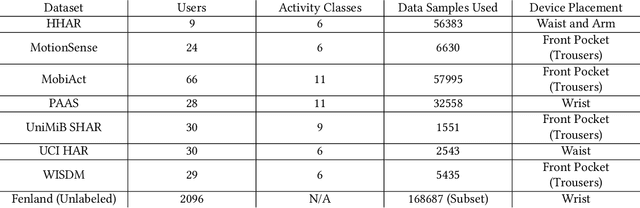
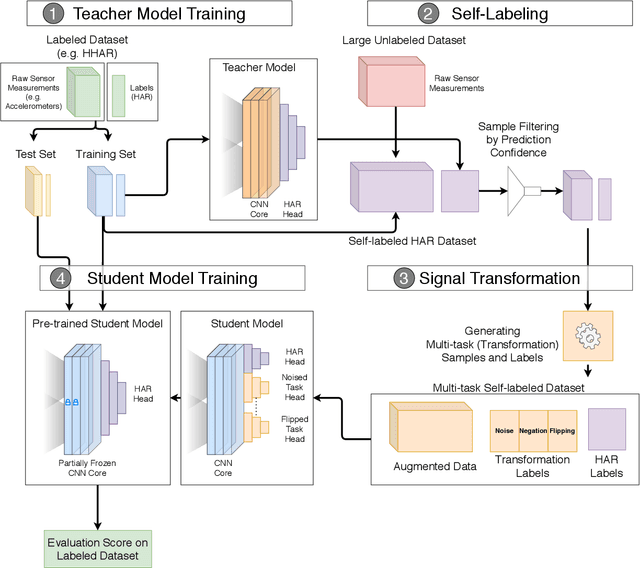
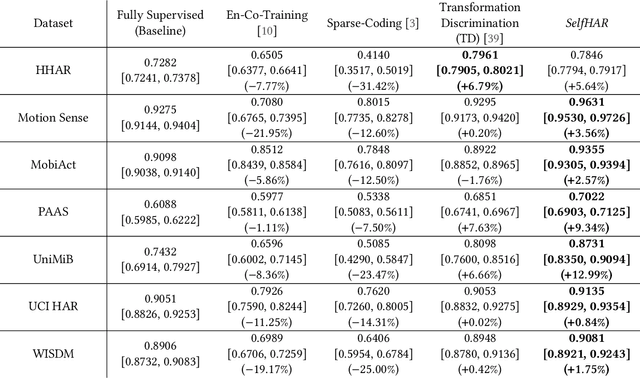
Abstract:Machine learning and deep learning have shown great promise in mobile sensing applications, including Human Activity Recognition. However, the performance of such models in real-world settings largely depends on the availability of large datasets that captures diverse behaviors. Recently, studies in computer vision and natural language processing have shown that leveraging massive amounts of unlabeled data enables performance on par with state-of-the-art supervised models. In this work, we present SelfHAR, a semi-supervised model that effectively learns to leverage unlabeled mobile sensing datasets to complement small labeled datasets. Our approach combines teacher-student self-training, which distills the knowledge of unlabeled and labeled datasets while allowing for data augmentation, and multi-task self-supervision, which learns robust signal-level representations by predicting distorted versions of the input. We evaluated SelfHAR on various HAR datasets and showed state-of-the-art performance over supervised and previous semi-supervised approaches, with up to 12% increase in F1 score using the same number of model parameters at inference. Furthermore, SelfHAR is data-efficient, reaching similar performance using up to 10 times less labeled data compared to supervised approaches. Our work not only achieves state-of-the-art performance in a diverse set of HAR datasets, but also sheds light on how pre-training tasks may affect downstream performance.
Self-supervised transfer learning of physiological representations from free-living wearable data
Nov 18, 2020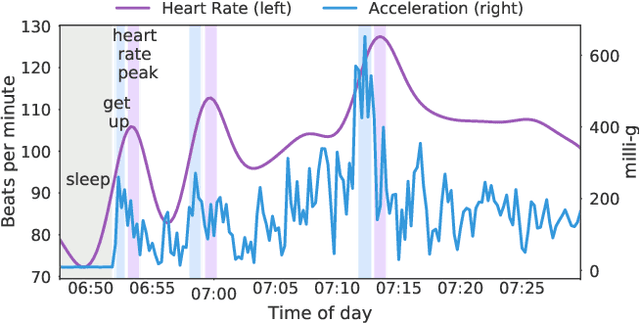
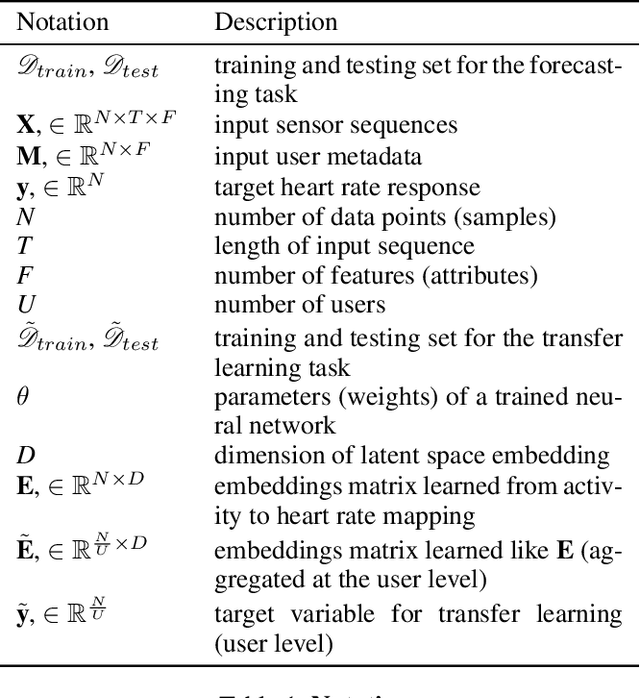
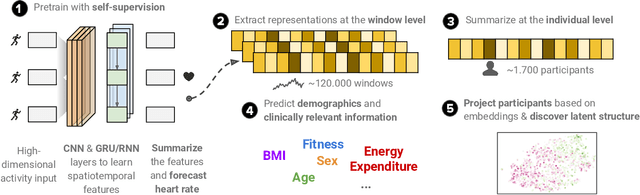
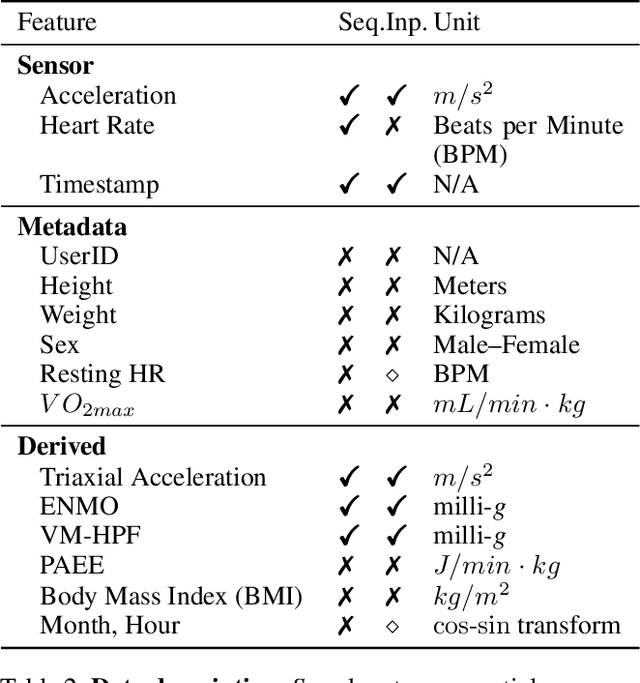
Abstract:Wearable devices such as smartwatches are becoming increasingly popular tools for objectively monitoring physical activity in free-living conditions. To date, research has primarily focused on the purely supervised task of human activity recognition, demonstrating limited success in inferring high-level health outcomes from low-level signals. Here, we present a novel self-supervised representation learning method using activity and heart rate (HR) signals without semantic labels. With a deep neural network, we set HR responses as the supervisory signal for the activity data, leveraging their underlying physiological relationship. In addition, we propose a custom quantile loss function that accounts for the long-tailed HR distribution present in the general population. We evaluate our model in the largest free-living combined-sensing dataset (comprising >280k hours of wrist accelerometer & wearable ECG data). Our contributions are two-fold: i) the pre-training task creates a model that can accurately forecast HR based only on cheap activity sensors, and ii) we leverage the information captured through this task by proposing a simple method to aggregate the learnt latent representations (embeddings) from the window-level to user-level. Notably, we show that the embeddings can generalize in various downstream tasks through transfer learning with linear classifiers, capturing physiologically meaningful, personalized information. For instance, they can be used to predict variables associated with individuals' health, fitness and demographic characteristics, outperforming unsupervised autoencoders and common bio-markers. Overall, we propose the first multimodal self-supervised method for behavioral and physiological data with implications for large-scale health and lifestyle monitoring.
Learning Generalizable Physiological Representations from Large-scale Wearable Data
Nov 09, 2020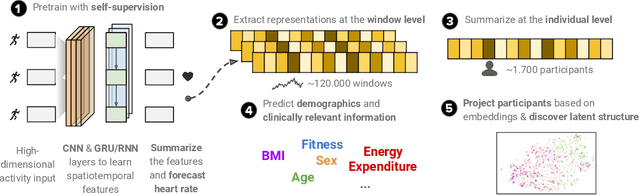
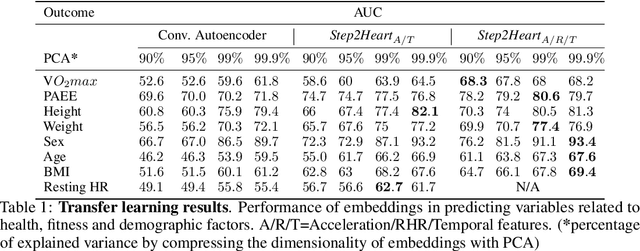
Abstract:To date, research on sensor-equipped mobile devices has primarily focused on the purely supervised task of human activity recognition (walking, running, etc), demonstrating limited success in inferring high-level health outcomes from low-level signals, such as acceleration. Here, we present a novel self-supervised representation learning method using activity and heart rate (HR) signals without semantic labels. With a deep neural network, we set HR responses as the supervisory signal for the activity data, leveraging their underlying physiological relationship. We evaluate our model in the largest free-living combined-sensing dataset (comprising more than 280,000 hours of wrist accelerometer & wearable ECG data) and show that the resulting embeddings can generalize in various downstream tasks through transfer learning with linear classifiers, capturing physiologically meaningful, personalized information. For instance, they can be used to predict (higher than 70 AUC) variables associated with individuals' health, fitness and demographic characteristics, outperforming unsupervised autoencoders and common bio-markers. Overall, we propose the first multimodal self-supervised method for behavioral and physiological data with implications for large-scale health and lifestyle monitoring.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge