Simona Cocco
LPENS, PSL
Restoring balance: principled under/oversampling of data for optimal classification
May 15, 2024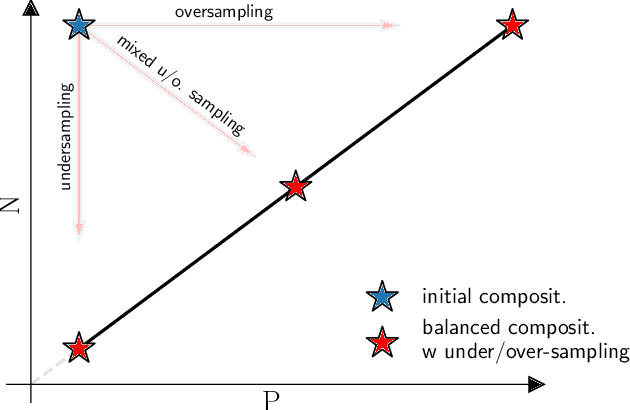
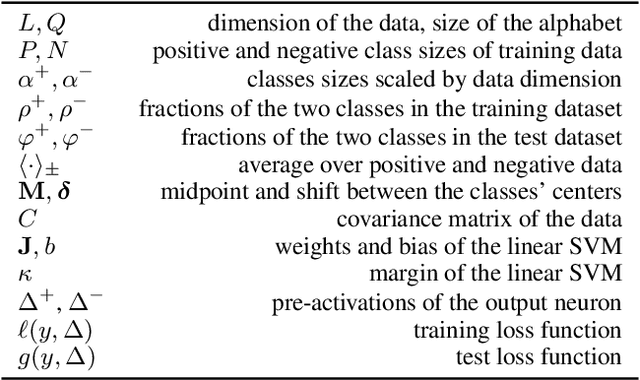
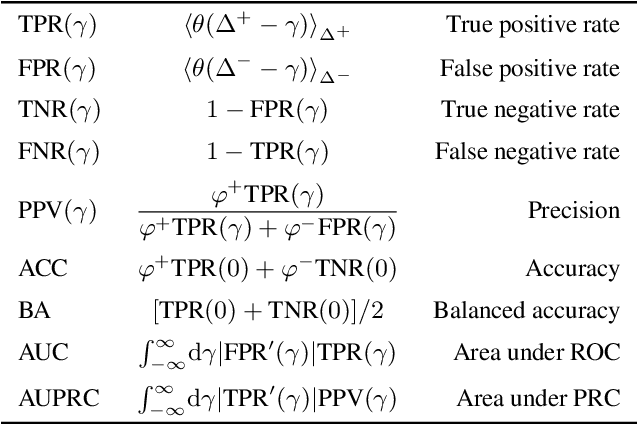
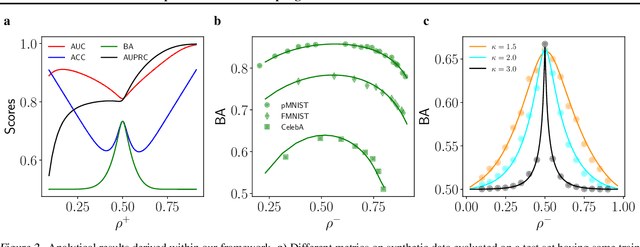
Abstract:Class imbalance in real-world data poses a common bottleneck for machine learning tasks, since achieving good generalization on under-represented examples is often challenging. Mitigation strategies, such as under or oversampling the data depending on their abundances, are routinely proposed and tested empirically, but how they should adapt to the data statistics remains poorly understood. In this work, we determine exact analytical expressions of the generalization curves in the high-dimensional regime for linear classifiers (Support Vector Machines). We also provide a sharp prediction of the effects of under/oversampling strategies depending on class imbalance, first and second moments of the data, and the metrics of performance considered. We show that mixed strategies involving under and oversampling of data lead to performance improvement. Through numerical experiments, we show the relevance of our theoretical predictions on real datasets, on deeper architectures and with sampling strategies based on unsupervised probabilistic models.
Disentangling representations in Restricted Boltzmann Machines without adversaries
Jul 01, 2022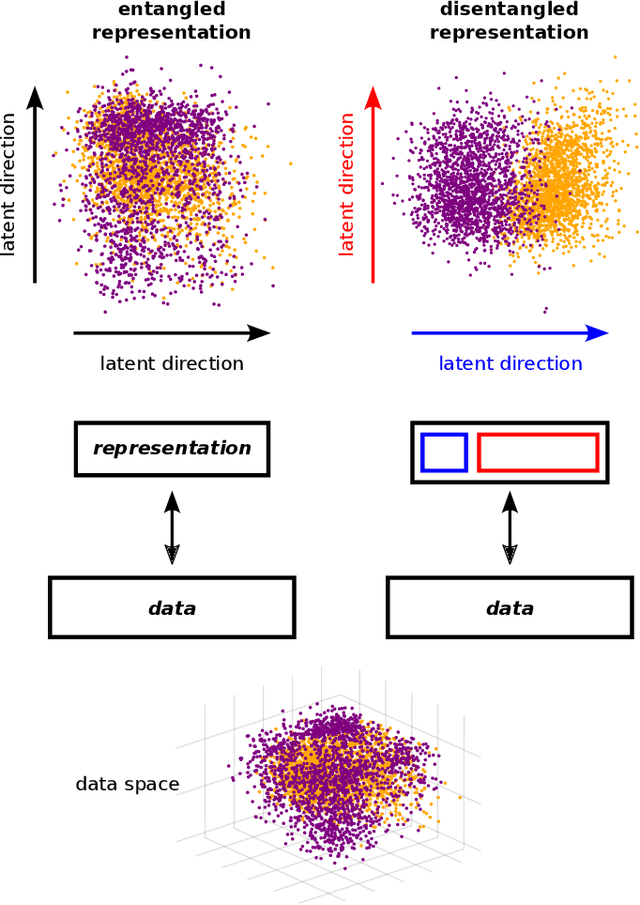
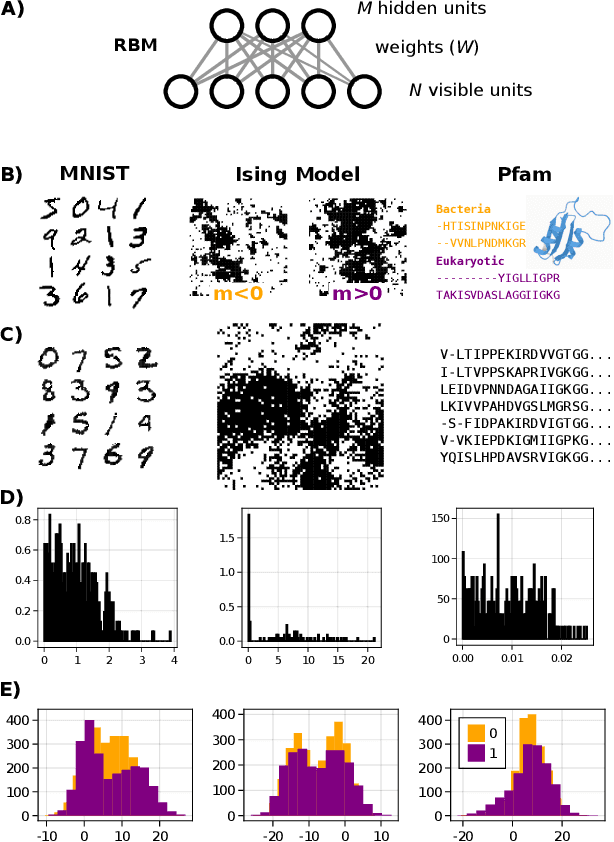
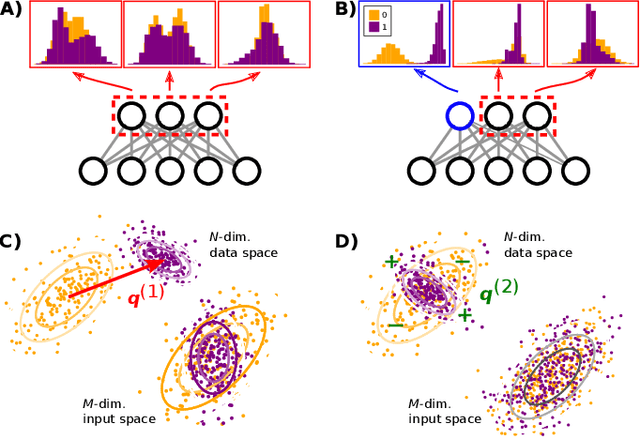
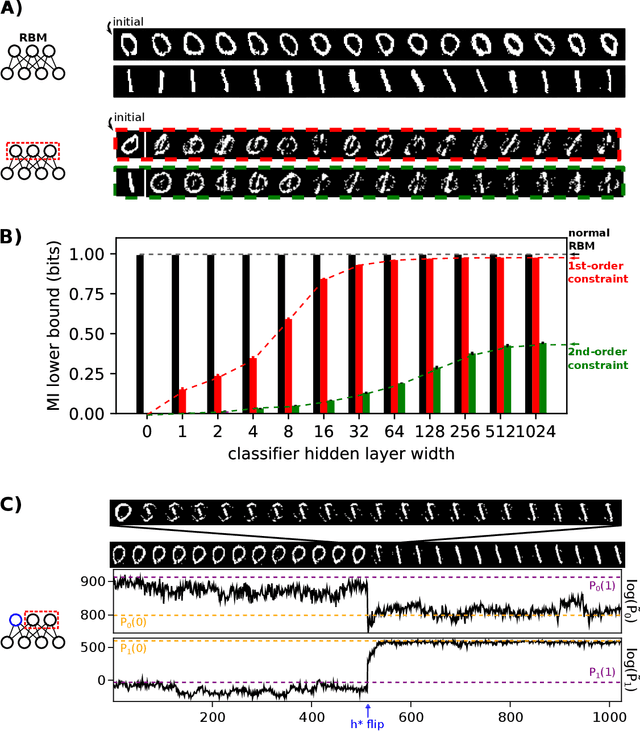
Abstract:A goal of unsupervised machine learning is to disentangle representations of complex high-dimensional data, allowing for interpreting the significant latent factors of variation in the data as well as for manipulating them to generate new data with desirable features. These methods often rely on an adversarial scheme, in which representations are tuned to avoid discriminators from being able to reconstruct specific data information (labels). We propose a simple, effective way of disentangling representations without any need to train adversarial discriminators, and apply our approach to Restricted Boltzmann Machines (RBM), one of the simplest representation-based generative models. Our approach relies on the introduction of adequate constraints on the weights during training, which allows us to concentrate information about labels on a small subset of latent variables. The effectiveness of the approach is illustrated on the MNIST dataset, the two-dimensional Ising model, and taxonomy of protein families. In addition, we show how our framework allows for computing the cost, in terms of log-likelihood of the data, associated to the disentanglement of their representations.
'Place-cell' emergence and learning of invariant data with restricted Boltzmann machines: breaking and dynamical restoration of continuous symmetries in the weight space
Dec 30, 2019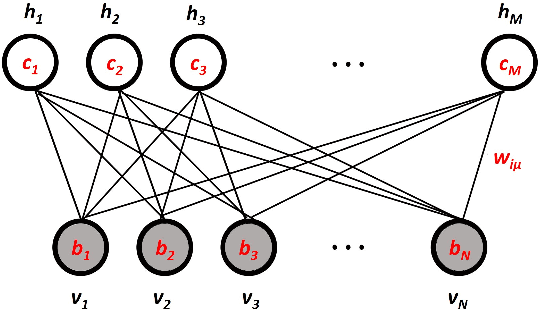


Abstract:Distributions of data or sensory stimuli often enjoy underlying invariances. How and to what extent those symmetries are captured by unsupervised learning methods is a relevant question in machine learning and in computational neuroscience. We study here, through a combination of numerical and analytical tools, the learning dynamics of Restricted Boltzmann Machines (RBM), a neural network paradigm for representation learning. As learning proceeds from a random configuration of the network weights, we show the existence of, and characterize a symmetry-breaking phenomenon, in which the latent variables acquire receptive fields focusing on limited parts of the invariant manifold supporting the data. The symmetry is restored at large learning times through the diffusion of the receptive field over the invariant manifold; hence, the RBM effectively spans a continuous attractor in the space of network weights. This symmetry-breaking phenomenon takes place only if the amount of data available for training exceeds some critical value, depending on the network size and the intensity of symmetry-induced correlations in the data; below this 'retarded-learning' threshold, the network weights are essentially noisy and overfit the data.
Learning Compositional Representations of Interacting Systems with Restricted Boltzmann Machines: Comparative Study of Lattice Proteins
Feb 18, 2019Abstract:A Restricted Boltzmann Machine (RBM) is an unsupervised machine-learning bipartite graphical model that jointly learns a probability distribution over data and extracts their relevant statistical features. As such, RBM were recently proposed for characterizing the patterns of coevolution between amino acids in protein sequences and for designing new sequences. Here, we study how the nature of the features learned by RBM changes with its defining parameters, such as the dimensionality of the representations (size of the hidden layer) and the sparsity of the features. We show that for adequate values of these parameters, RBM operate in a so-called compositional phase in which visible configurations sampled from the RBM are obtained by recombining these features. We then compare the performance of RBM with other standard representation learning algorithms, including Principal or Independent Component Analysis, autoencoders (AE), variational auto-encoders (VAE), and their sparse variants. We show that RBM, due to the stochastic mapping between data configurations and representations, better capture the underlying interactions in the system and are significantly more robust with respect to sample size than deterministic methods such as PCA or ICA. In addition, this stochastic mapping is not prescribed a priori as in VAE, but learned from data, which allows RBM to show good performance even with shallow architectures. All numerical results are illustrated on synthetic lattice-protein data, that share similar statistical features with real protein sequences, and for which ground-truth interactions are known.
High-Dimensional Inference with the generalized Hopfield Model: Principal Component Analysis and Corrections
Apr 19, 2011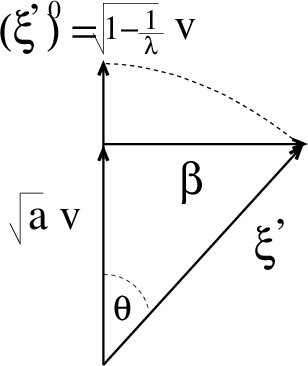
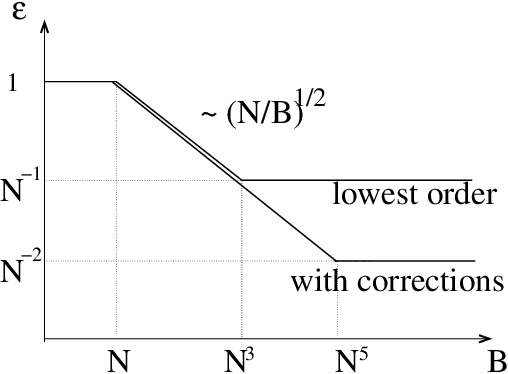
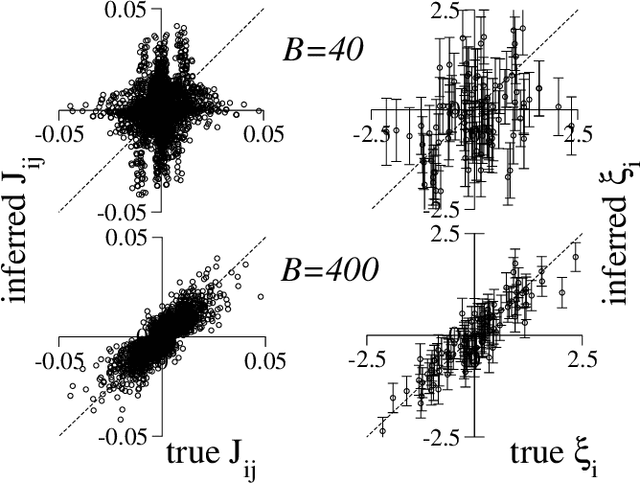
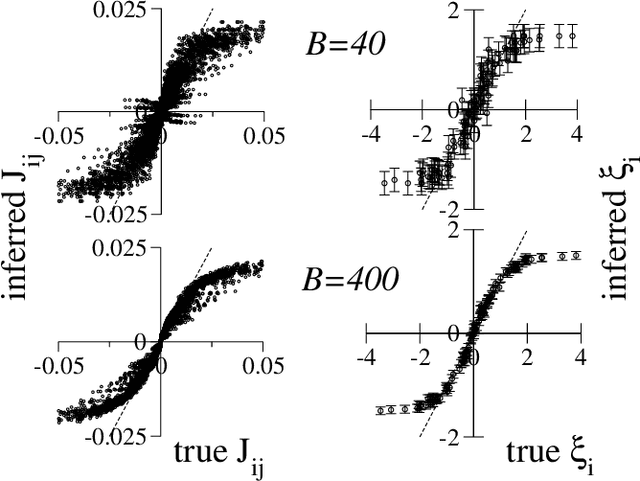
Abstract:We consider the problem of inferring the interactions between a set of N binary variables from the knowledge of their frequencies and pairwise correlations. The inference framework is based on the Hopfield model, a special case of the Ising model where the interaction matrix is defined through a set of patterns in the variable space, and is of rank much smaller than N. We show that Maximum Lik elihood inference is deeply related to Principal Component Analysis when the amp litude of the pattern components, xi, is negligible compared to N^1/2. Using techniques from statistical mechanics, we calculate the corrections to the patterns to the first order in xi/N^1/2. We stress that it is important to generalize the Hopfield model and include both attractive and repulsive patterns, to correctly infer networks with sparse and strong interactions. We present a simple geometrical criterion to decide how many attractive and repulsive patterns should be considered as a function of the sampling noise. We moreover discuss how many sampled configurations are required for a good inference, as a function of the system size, N and of the amplitude, xi. The inference approach is illustrated on synthetic and biological data.
Fast Inference of Interactions in Assemblies of Stochastic Integrate-and-Fire Neurons from Spike Recordings
Feb 25, 2011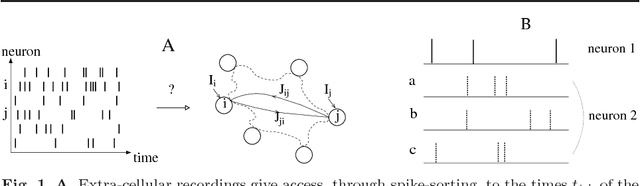
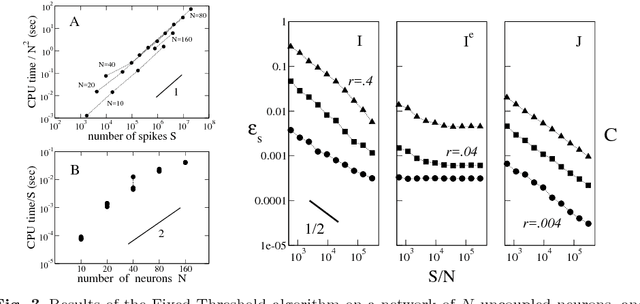
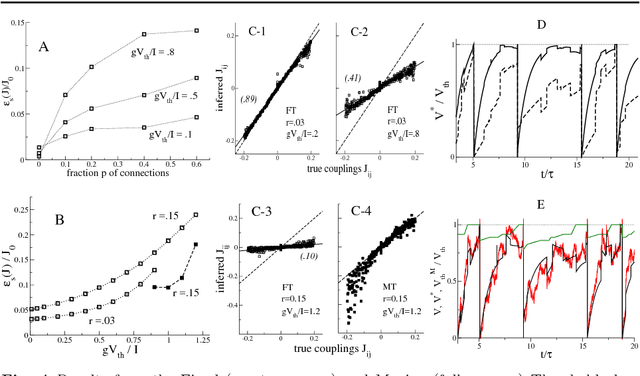
Abstract:We present two Bayesian procedures to infer the interactions and external currents in an assembly of stochastic integrate-and-fire neurons from the recording of their spiking activity. The first procedure is based on the exact calculation of the most likely time courses of the neuron membrane potentials conditioned by the recorded spikes, and is exact for a vanishing noise variance and for an instantaneous synaptic integration. The second procedure takes into account the presence of fluctuations around the most likely time courses of the potentials, and can deal with moderate noise levels. The running time of both procedures is proportional to the number S of spikes multiplied by the squared number N of neurons. The algorithms are validated on synthetic data generated by networks with known couplings and currents. We also reanalyze previously published recordings of the activity of the salamander retina (including from 32 to 40 neurons, and from 65,000 to 170,000 spikes). We study the dependence of the inferred interactions on the membrane leaking time; the differences and similarities with the classical cross-correlation analysis are discussed.
Adaptive Cluster Expansion for Inferring Boltzmann Machines with Noisy Data
Feb 16, 2011



Abstract:We introduce a procedure to infer the interactions among a set of binary variables, based on their sampled frequencies and pairwise correlations. The algorithm builds the clusters of variables contributing most to the entropy of the inferred Ising model, and rejects the small contributions due to the sampling noise. Our procedure successfully recovers benchmark Ising models even at criticality and in the low temperature phase, and is applied to neurobiological data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge