Sean P. Fitzgibbon
ICAM-reg: Interpretable Classification and Regression with Feature Attribution for Mapping Neurological Phenotypes in Individual Scans
Mar 03, 2021



Abstract:An important goal of medical imaging is to be able to precisely detect patterns of disease specific to individual scans; however, this is challenged in brain imaging by the degree of heterogeneity of shape and appearance. Traditional methods, based on image registration to a global template, historically fail to detect variable features of disease, as they utilise population-based analyses, suited primarily to studying group-average effects. In this paper we therefore take advantage of recent developments in generative deep learning to develop a method for simultaneous classification, or regression, and feature attribution (FA). Specifically, we explore the use of a VAE-GAN translation network called ICAM, to explicitly disentangle class relevant features from background confounds for improved interpretability and regression of neurological phenotypes. We validate our method on the tasks of Mini-Mental State Examination (MMSE) cognitive test score prediction for the Alzheimer's Disease Neuroimaging Initiative (ADNI) cohort, as well as brain age prediction, for both neurodevelopment and neurodegeneration, using the developing Human Connectome Project (dHCP) and UK Biobank datasets. We show that the generated FA maps can be used to explain outlier predictions and demonstrate that the inclusion of a regression module improves the disentanglement of the latent space. Our code is freely available on Github https://github.com/CherBass/ICAM.
Reducing training requirements through evolutionary based dimension reduction and subject transfer
Feb 06, 2016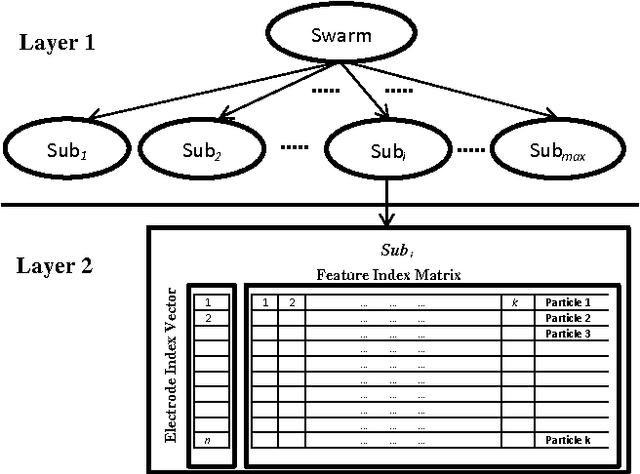
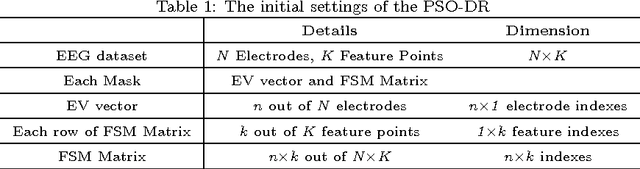

Abstract:Training Brain Computer Interface (BCI) systems to understand the intention of a subject through Electroencephalogram (EEG) data currently requires multiple training sessions with a subject in order to develop the necessary expertise to distinguish signals for different tasks. Conventionally the task of training the subject is done by introducing a training and calibration stage during which some feedback is presented to the subject. This training session can take several hours which is not appropriate for on-line EEG-based BCI systems. An alternative approach is to use previous recording sessions of the same person or some other subjects that performed the same tasks (subject transfer) for training the classifiers. The main aim of this study is to generate a methodology that allows the use of data from other subjects while reducing the dimensions of the data. The study investigates several possibilities for reducing the necessary training and calibration period in subjects and the classifiers and addresses the impact of i) evolutionary subject transfer and ii) adapting previously trained methods (retraining) using other subjects data. Our results suggest reduction to 40% of target subject data is sufficient for training the classifier. Our results also indicate the superiority of the approaches that incorporated evolutionary subject transfer and highlights the feasibility of adapting a system trained on other subjects.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge