Sara Silva
Evolving Financial Trading Strategies with Vectorial Genetic Programming
Apr 07, 2025



Abstract:Establishing profitable trading strategies in financial markets is a challenging task. While traditional methods like technical analysis have long served as foundational tools for traders to recognize and act upon market patterns, the evolving landscape has called for more advanced techniques. We explore the use of Vectorial Genetic Programming (VGP) for this task, introducing two new variants of VGP, one that allows operations with complex numbers and another that implements a strongly-typed version of VGP. We evaluate the different variants on three financial instruments, with datasets spanning more than seven years. Despite the inherent difficulty of this task, it was possible to evolve profitable trading strategies. A comparative analysis of the three VGP variants and standard GP revealed that standard GP is always among the worst whereas strongly-typed VGP is always among the best.
Graph Contrastive Learning for Connectome Classification
Feb 07, 2025Abstract:With recent advancements in non-invasive techniques for measuring brain activity, such as magnetic resonance imaging (MRI), the study of structural and functional brain networks through graph signal processing (GSP) has gained notable prominence. GSP stands as a key tool in unraveling the interplay between the brain's function and structure, enabling the analysis of graphs defined by the connections between regions of interest -- referred to as connectomes in this context. Our work represents a further step in this direction by exploring supervised contrastive learning methods within the realm of graph representation learning. The main objective of this approach is to generate subject-level (i.e., graph-level) vector representations that bring together subjects sharing the same label while separating those with different labels. These connectome embeddings are derived from a graph neural network Encoder-Decoder architecture, which jointly considers structural and functional connectivity. By leveraging data augmentation techniques, the proposed framework achieves state-of-the-art performance in a gender classification task using Human Connectome Project data. More broadly, our connectome-centric methodological advances support the promising prospect of using GSP to discover more about brain function, with potential impact to understanding heterogeneity in the neurodegeneration for precision medicine and diagnosis.
Testing the Segment Anything Model on radiology data
Dec 20, 2023



Abstract:Deep learning models trained with large amounts of data have become a recent and effective approach to predictive problem solving -- these have become known as "foundation models" as they can be used as fundamental tools for other applications. While the paramount examples of image classification (earlier) and large language models (more recently) led the way, the Segment Anything Model (SAM) was recently proposed and stands as the first foundation model for image segmentation, trained on over 10 million images and with recourse to over 1 billion masks. However, the question remains -- what are the limits of this foundation? Given that magnetic resonance imaging (MRI) stands as an important method of diagnosis, we sought to understand whether SAM could be used for a few tasks of zero-shot segmentation using MRI data. Particularly, we wanted to know if selecting masks from the pool of SAM predictions could lead to good segmentations. Here, we provide a critical assessment of the performance of SAM on magnetic resonance imaging data. We show that, while acceptable in a very limited set of cases, the overall trend implies that these models are insufficient for MRI segmentation across the whole volume, but can provide good segmentations in a few, specific slices. More importantly, we note that while foundation models trained on natural images are set to become key aspects of predictive modelling, they may prove ineffective when used on other imaging modalities.
Biomedical Knowledge Graph Embeddings with Negative Statements
Aug 07, 2023



Abstract:A knowledge graph is a powerful representation of real-world entities and their relations. The vast majority of these relations are defined as positive statements, but the importance of negative statements is increasingly recognized, especially under an Open World Assumption. Explicitly considering negative statements has been shown to improve performance on tasks such as entity summarization and question answering or domain-specific tasks such as protein function prediction. However, no attention has been given to the exploration of negative statements by knowledge graph embedding approaches despite the potential of negative statements to produce more accurate representations of entities in a knowledge graph. We propose a novel approach, TrueWalks, to incorporate negative statements into the knowledge graph representation learning process. In particular, we present a novel walk-generation method that is able to not only differentiate between positive and negative statements but also take into account the semantic implications of negation in ontology-rich knowledge graphs. This is of particular importance for applications in the biomedical domain, where the inadequacy of embedding approaches regarding negative statements at the ontology level has been identified as a crucial limitation. We evaluate TrueWalks in ontology-rich biomedical knowledge graphs in two different predictive tasks based on KG embeddings: protein-protein interaction prediction and gene-disease association prediction. We conduct an extensive analysis over established benchmarks and demonstrate that our method is able to improve the performance of knowledge graph embeddings on all tasks.
Benchmark datasets for biomedical knowledge graphs with negative statements
Jul 21, 2023



Abstract:Knowledge graphs represent facts about real-world entities. Most of these facts are defined as positive statements. The negative statements are scarce but highly relevant under the open-world assumption. Furthermore, they have been demonstrated to improve the performance of several applications, namely in the biomedical domain. However, no benchmark dataset supports the evaluation of the methods that consider these negative statements. We present a collection of datasets for three relation prediction tasks - protein-protein interaction prediction, gene-disease association prediction and disease prediction - that aim at circumventing the difficulties in building benchmarks for knowledge graphs with negative statements. These datasets include data from two successful biomedical ontologies, Gene Ontology and Human Phenotype Ontology, enriched with negative statements. We also generate knowledge graph embeddings for each dataset with two popular path-based methods and evaluate the performance in each task. The results show that the negative statements can improve the performance of knowledge graph embeddings.
Explainable Representations for Relation Prediction in Knowledge Graphs
Jun 22, 2023



Abstract:Knowledge graphs represent real-world entities and their relations in a semantically-rich structure supported by ontologies. Exploring this data with machine learning methods often relies on knowledge graph embeddings, which produce latent representations of entities that preserve structural and local graph neighbourhood properties, but sacrifice explainability. However, in tasks such as link or relation prediction, understanding which specific features better explain a relation is crucial to support complex or critical applications. We propose SEEK, a novel approach for explainable representations to support relation prediction in knowledge graphs. It is based on identifying relevant shared semantic aspects (i.e., subgraphs) between entities and learning representations for each subgraph, producing a multi-faceted and explainable representation. We evaluate SEEK on two real-world highly complex relation prediction tasks: protein-protein interaction prediction and gene-disease association prediction. Our extensive analysis using established benchmarks demonstrates that SEEK achieves significantly better performance than standard learning representation methods while identifying both sufficient and necessary explanations based on shared semantic aspects.
Plotting time: On the usage of CNNs for time series classification
Feb 08, 2021



Abstract:We present a novel approach for time series classification where we represent time series data as plot images and feed them to a simple CNN, outperforming several state-of-the-art methods. We propose a simple and highly replicable way of plotting the time series, and feed these images as input to a non-optimized shallow CNN, without any normalization or residual connections. These representations are no more than default line plots using the time series data, where the only pre-processing applied is to reduce the number of white pixels in the image. We compare our method with different state-of-the-art methods specialized in time series classification on two real-world non public datasets, as well as 98 datasets of the UCR dataset collection. The results show that our approach is very promising, achieving the best results on both real-world datasets and matching / beating the best state-of-the-art methods in six UCR datasets. We argue that, if a simple naive design like ours can obtain such good results, it is worth further exploring the capabilities of using image representation of time series data, along with more powerful CNNs, for classification and other related tasks.
Improving the Detection of Burnt Areas in Remote Sensing using Hyper-features Evolved by M3GP
Jan 31, 2020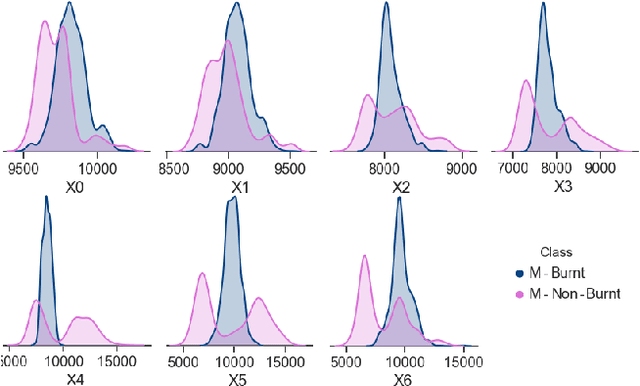
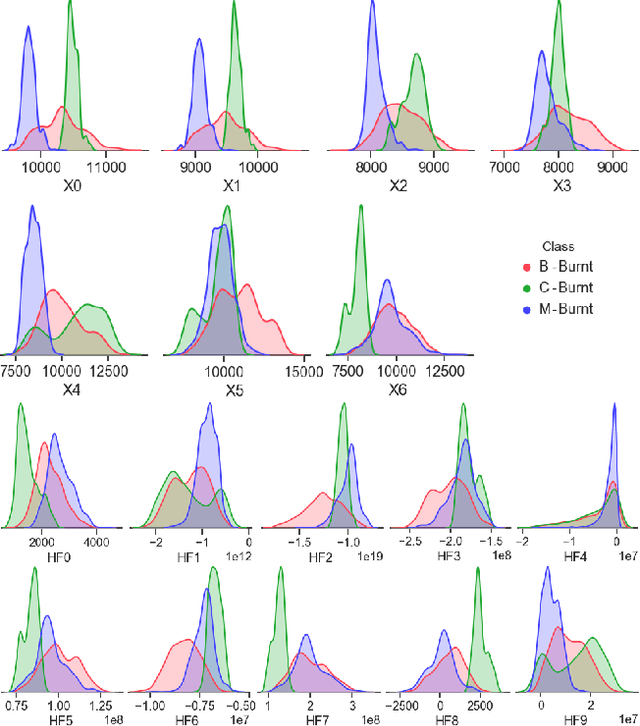
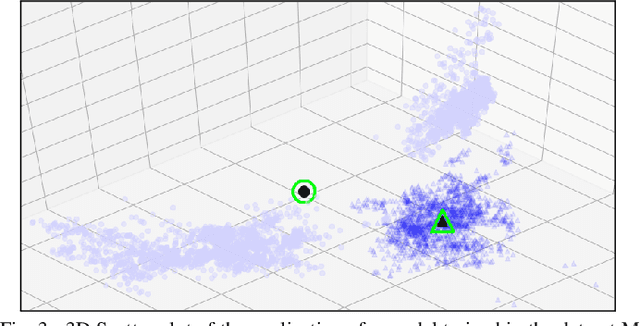
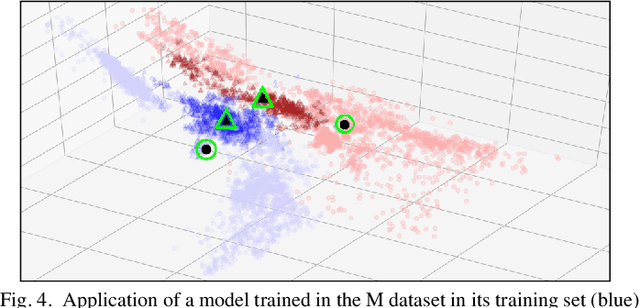
Abstract:One problem found when working with satellite images is the radiometric variations across the image and different images. Intending to improve remote sensing models for the classification of burnt areas, we set two objectives. The first is to understand the relationship between feature spaces and the predictive ability of the models, allowing us to explain the differences between learning and generalization when training and testing in different datasets. We find that training on datasets built from more than one image provides models that generalize better. These results are explained by visualizing the dispersion of values on the feature space. The second objective is to evolve hyper-features that improve the performance of different classifiers on a variety of test sets. We find the hyper-features to be beneficial, and obtain the best models with XGBoost, even if the hyper-features are optimized for a different method.
A Study of Fitness Landscapes for Neuroevolution
Jan 30, 2020

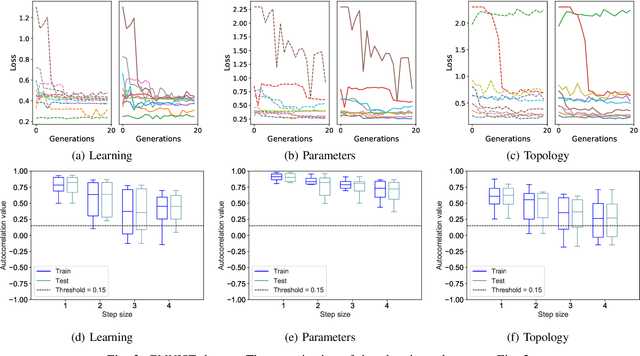
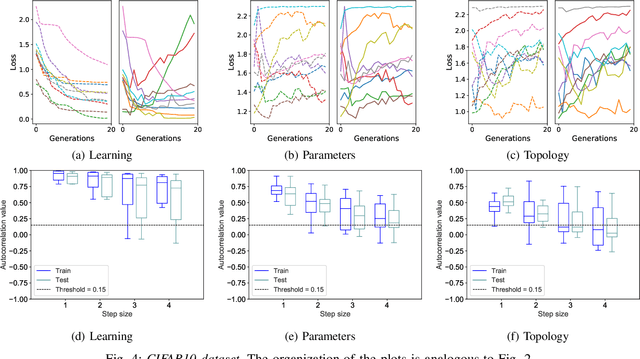
Abstract:Fitness landscapes are a useful concept to study the dynamics of meta-heuristics. In the last two decades, they have been applied with success to estimate the optimization power of several types of evolutionary algorithms, including genetic algorithms and genetic programming. However, so far they have never been used to study the performance of machine learning algorithms on unseen data, and they have never been applied to neuroevolution. This paper aims at filling both these gaps, applying for the first time fitness landscapes to neuroevolution and using them to infer useful information about the predictive ability of the method. More specifically, we use a grammar-based approach to generate convolutional neural networks, and we study the dynamics of three different mutations to evolve them. To characterize fitness landscapes, we study autocorrelation and entropic measure of ruggedness. The results show that these measures are appropriate for estimating both the optimization power and the generalization ability of the considered neuroevolution configurations.
Ensemble Genetic Programming
Jan 21, 2020



Abstract:Ensemble learning is a powerful paradigm that has been usedin the top state-of-the-art machine learning methods like Random Forestsand XGBoost. Inspired by the success of such methods, we have devel-oped a new Genetic Programming method called Ensemble GP. The evo-lutionary cycle of Ensemble GP follows the same steps as other GeneticProgramming systems, but with differences in the population structure,fitness evaluation and genetic operators. We have tested this method oneight binary classification problems, achieving results significantly betterthan standard GP, with much smaller models. Although other methodslike M3GP and XGBoost were the best overall, Ensemble GP was able toachieve exceptionally good generalization results on a particularly hardproblem where none of the other methods was able to succeed.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge