Nuno M. Rodrigues
Testing the Segment Anything Model on radiology data
Dec 20, 2023



Abstract:Deep learning models trained with large amounts of data have become a recent and effective approach to predictive problem solving -- these have become known as "foundation models" as they can be used as fundamental tools for other applications. While the paramount examples of image classification (earlier) and large language models (more recently) led the way, the Segment Anything Model (SAM) was recently proposed and stands as the first foundation model for image segmentation, trained on over 10 million images and with recourse to over 1 billion masks. However, the question remains -- what are the limits of this foundation? Given that magnetic resonance imaging (MRI) stands as an important method of diagnosis, we sought to understand whether SAM could be used for a few tasks of zero-shot segmentation using MRI data. Particularly, we wanted to know if selecting masks from the pool of SAM predictions could lead to good segmentations. Here, we provide a critical assessment of the performance of SAM on magnetic resonance imaging data. We show that, while acceptable in a very limited set of cases, the overall trend implies that these models are insufficient for MRI segmentation across the whole volume, but can provide good segmentations in a few, specific slices. More importantly, we note that while foundation models trained on natural images are set to become key aspects of predictive modelling, they may prove ineffective when used on other imaging modalities.
Plotting time: On the usage of CNNs for time series classification
Feb 08, 2021



Abstract:We present a novel approach for time series classification where we represent time series data as plot images and feed them to a simple CNN, outperforming several state-of-the-art methods. We propose a simple and highly replicable way of plotting the time series, and feed these images as input to a non-optimized shallow CNN, without any normalization or residual connections. These representations are no more than default line plots using the time series data, where the only pre-processing applied is to reduce the number of white pixels in the image. We compare our method with different state-of-the-art methods specialized in time series classification on two real-world non public datasets, as well as 98 datasets of the UCR dataset collection. The results show that our approach is very promising, achieving the best results on both real-world datasets and matching / beating the best state-of-the-art methods in six UCR datasets. We argue that, if a simple naive design like ours can obtain such good results, it is worth further exploring the capabilities of using image representation of time series data, along with more powerful CNNs, for classification and other related tasks.
A Study of Fitness Landscapes for Neuroevolution
Jan 30, 2020

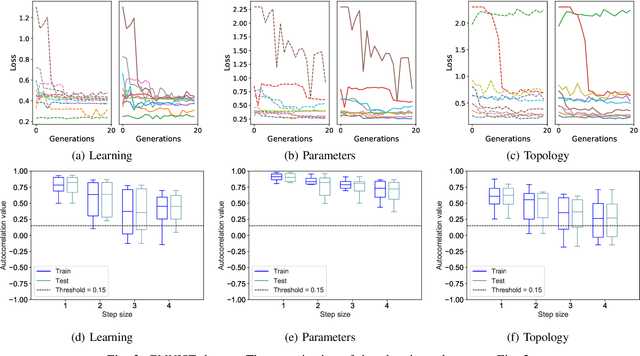
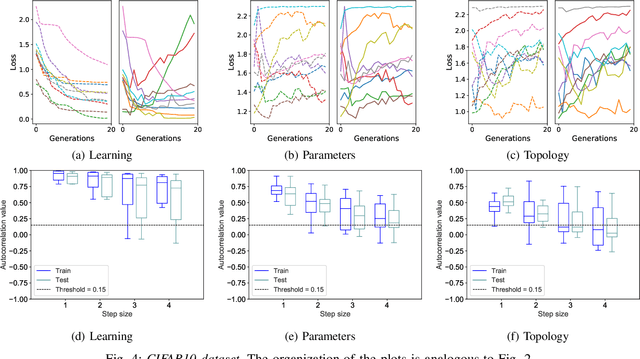
Abstract:Fitness landscapes are a useful concept to study the dynamics of meta-heuristics. In the last two decades, they have been applied with success to estimate the optimization power of several types of evolutionary algorithms, including genetic algorithms and genetic programming. However, so far they have never been used to study the performance of machine learning algorithms on unseen data, and they have never been applied to neuroevolution. This paper aims at filling both these gaps, applying for the first time fitness landscapes to neuroevolution and using them to infer useful information about the predictive ability of the method. More specifically, we use a grammar-based approach to generate convolutional neural networks, and we study the dynamics of three different mutations to evolve them. To characterize fitness landscapes, we study autocorrelation and entropic measure of ruggedness. The results show that these measures are appropriate for estimating both the optimization power and the generalization ability of the considered neuroevolution configurations.
Ensemble Genetic Programming
Jan 21, 2020



Abstract:Ensemble learning is a powerful paradigm that has been usedin the top state-of-the-art machine learning methods like Random Forestsand XGBoost. Inspired by the success of such methods, we have devel-oped a new Genetic Programming method called Ensemble GP. The evo-lutionary cycle of Ensemble GP follows the same steps as other GeneticProgramming systems, but with differences in the population structure,fitness evaluation and genetic operators. We have tested this method oneight binary classification problems, achieving results significantly betterthan standard GP, with much smaller models. Although other methodslike M3GP and XGBoost were the best overall, Ensemble GP was able toachieve exceptionally good generalization results on a particularly hardproblem where none of the other methods was able to succeed.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge