Santiago Fernandez Gonzalez
Cell Behavior Video Classification Challenge, a benchmark for computer vision methods in time-lapse microscopy
Jan 15, 2026Abstract:The classification of microscopy videos capturing complex cellular behaviors is crucial for understanding and quantifying the dynamics of biological processes over time. However, it remains a frontier in computer vision, requiring approaches that effectively model the shape and motion of objects without rigid boundaries, extract hierarchical spatiotemporal features from entire image sequences rather than static frames, and account for multiple objects within the field of view. To this end, we organized the Cell Behavior Video Classification Challenge (CBVCC), benchmarking 35 methods based on three approaches: classification of tracking-derived features, end-to-end deep learning architectures to directly learn spatiotemporal features from the entire video sequence without explicit cell tracking, or ensembling tracking-derived with image-derived features. We discuss the results achieved by the participants and compare the potential and limitations of each approach, serving as a basis to foster the development of computer vision methods for studying cellular dynamics.
A shortest-path based clustering algorithm for joint human-machine analysis of complex datasets
Dec 31, 2018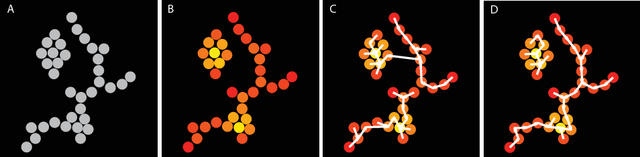
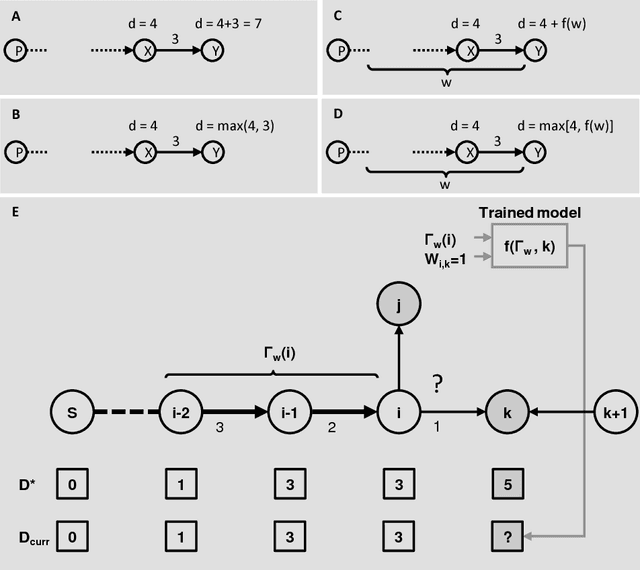
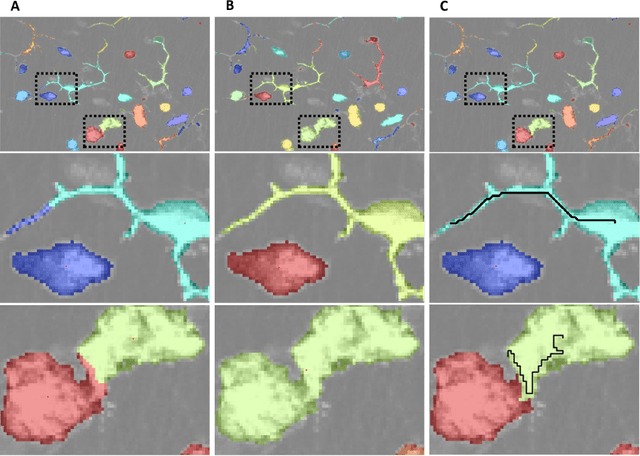
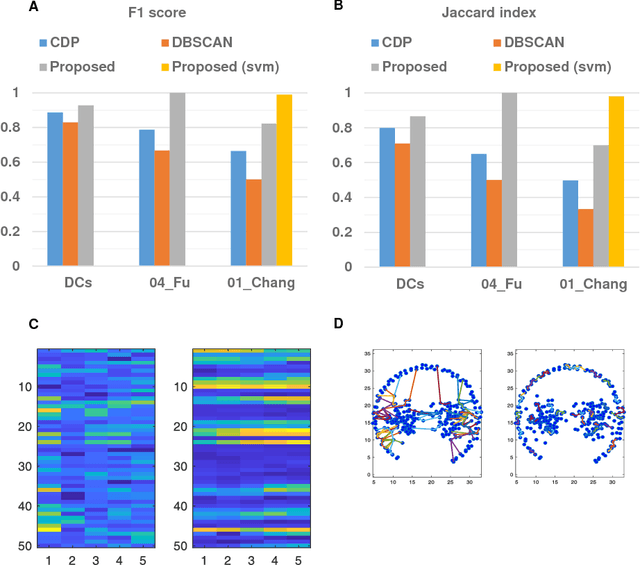
Abstract:Clustering is a technique for the analysis of datasets obtained by empirical studies in several disciplines with a major application for biomedical research. Essentially, clustering algorithms are executed by machines aiming at finding groups of related points in a dataset. However, the result of grouping depends on both metrics for point-to-point similarity and rules for point-to-group association. Indeed, non-appropriate metrics and rules can lead to undesirable clustering artifacts. This is especially relevant for datasets, where groups with heterogeneous structures co-exist. In this work, we propose an algorithm that achieves clustering by exploring the paths between points. This allows both, to evaluate the properties of the path (such as gaps, density variations, etc.), and expressing the preference for certain paths. Moreover, our algorithm supports the integration of existing knowledge about admissible and non-admissible clusters by training a path classifier. We demonstrate the accuracy of the proposed method on challenging datasets including points from synthetic shapes in publicly available benchmarks and microscopy data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge