A shortest-path based clustering algorithm for joint human-machine analysis of complex datasets
Paper and Code
Dec 31, 2018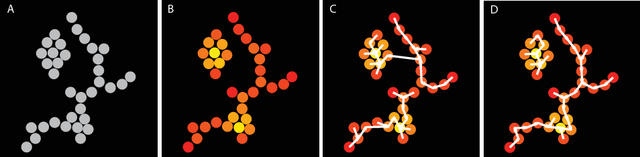
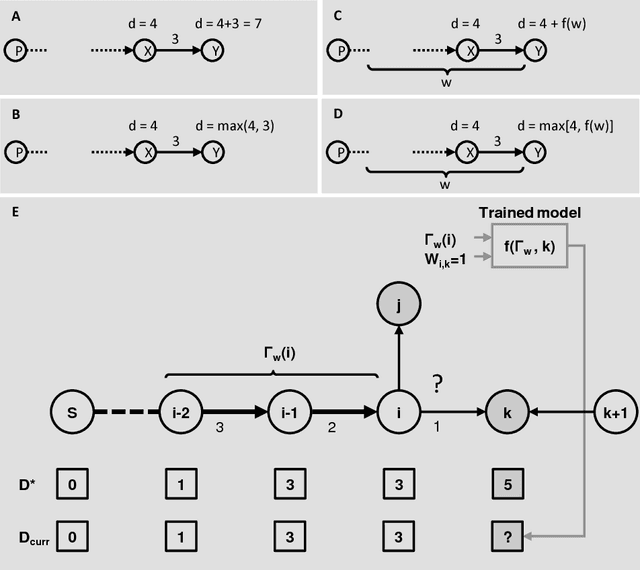
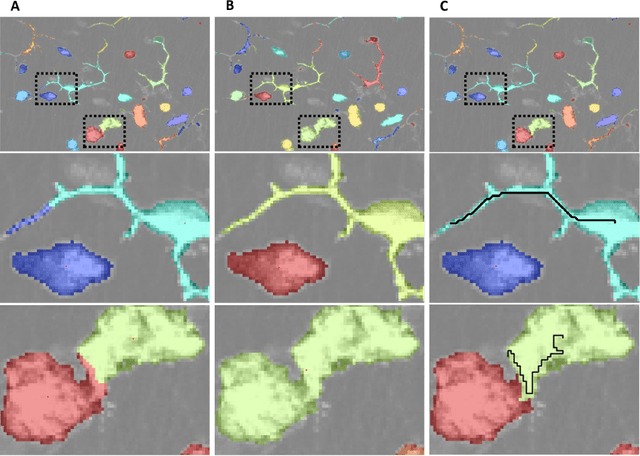
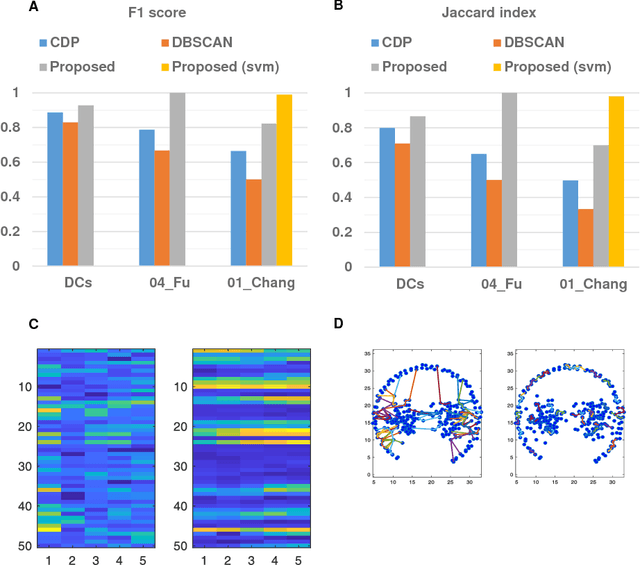
Clustering is a technique for the analysis of datasets obtained by empirical studies in several disciplines with a major application for biomedical research. Essentially, clustering algorithms are executed by machines aiming at finding groups of related points in a dataset. However, the result of grouping depends on both metrics for point-to-point similarity and rules for point-to-group association. Indeed, non-appropriate metrics and rules can lead to undesirable clustering artifacts. This is especially relevant for datasets, where groups with heterogeneous structures co-exist. In this work, we propose an algorithm that achieves clustering by exploring the paths between points. This allows both, to evaluate the properties of the path (such as gaps, density variations, etc.), and expressing the preference for certain paths. Moreover, our algorithm supports the integration of existing knowledge about admissible and non-admissible clusters by training a path classifier. We demonstrate the accuracy of the proposed method on challenging datasets including points from synthetic shapes in publicly available benchmarks and microscopy data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge