Sami Hamdan
for the Alzheimer's Disease Neuroimaging Initiative
Empirical Comparison between Cross-Validation and Mutation-Validation in Model Selection
Nov 23, 2023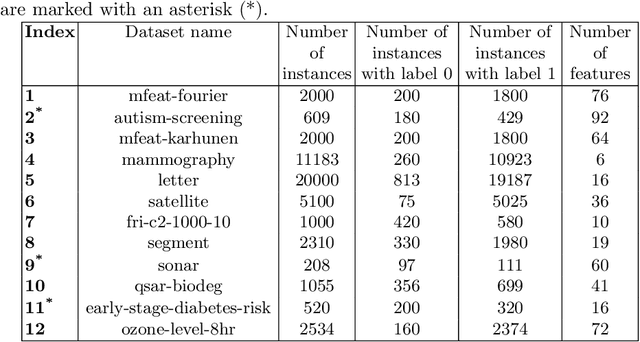
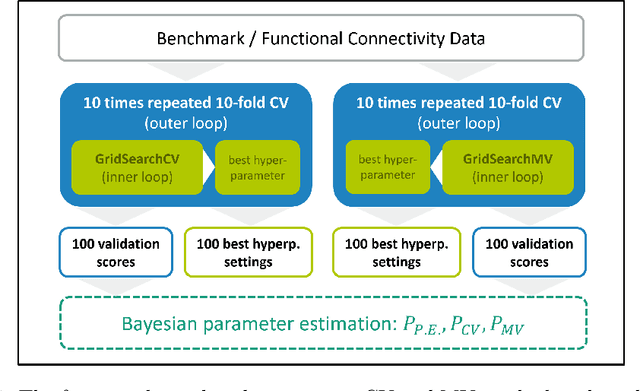
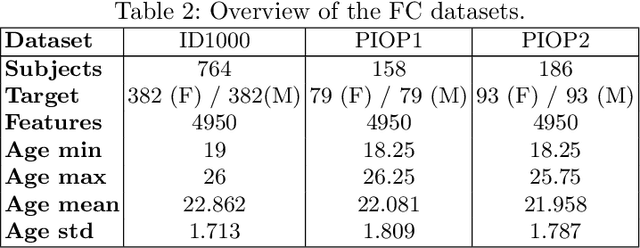
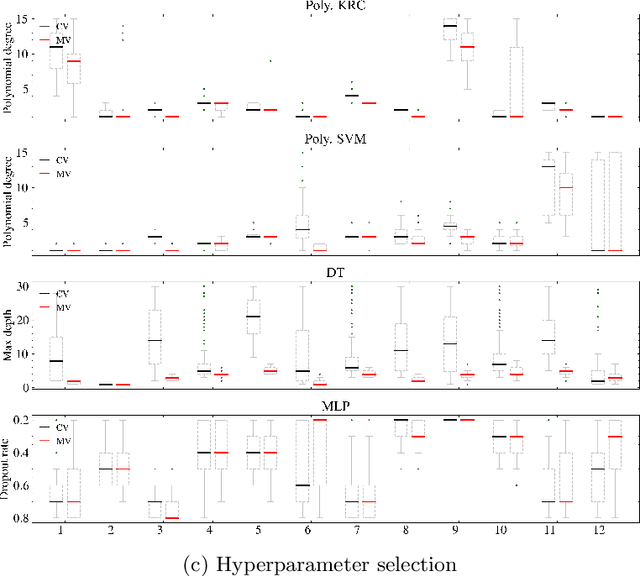
Abstract:Mutation validation (MV) is a recently proposed approach for model selection, garnering significant interest due to its unique characteristics and potential benefits compared to the widely used cross-validation (CV) method. In this study, we empirically compared MV and $k$-fold CV using benchmark and real-world datasets. By employing Bayesian tests, we compared generalization estimates yielding three posterior probabilities: practical equivalence, CV superiority, and MV superiority. We also evaluated the differences in the capacity of the selected models and computational efficiency. We found that both MV and CV select models with practically equivalent generalization performance across various machine learning algorithms and the majority of benchmark datasets. MV exhibited advantages in terms of selecting simpler models and lower computational costs. However, in some cases MV selected overly simplistic models leading to underfitting and showed instability in hyperparameter selection. These limitations of MV became more evident in the evaluation of a real-world neuroscientific task of predicting sex at birth using brain functional connectivity.
On Leakage in Machine Learning Pipelines
Nov 07, 2023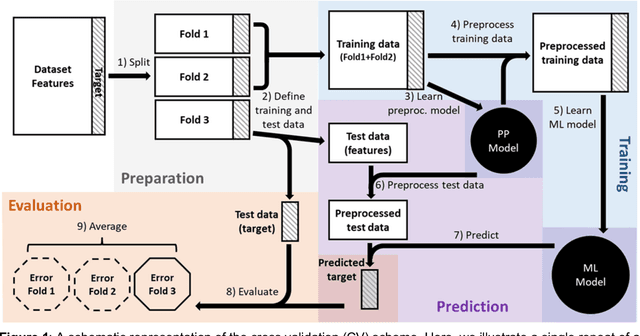
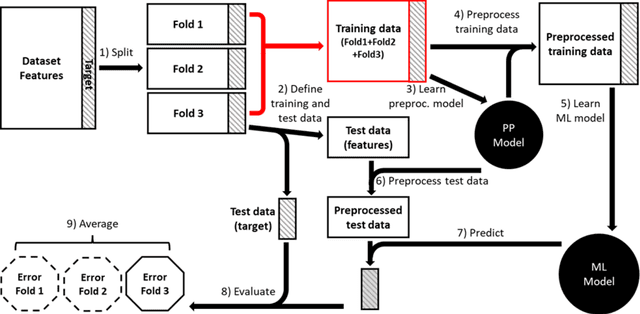
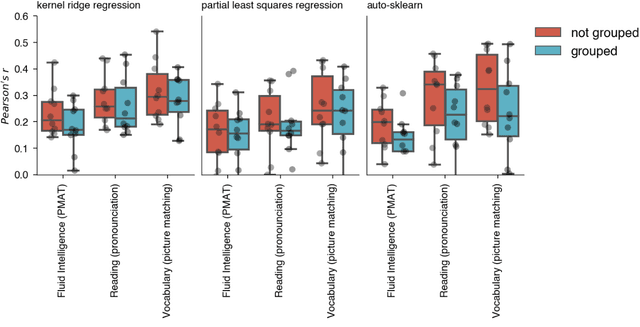
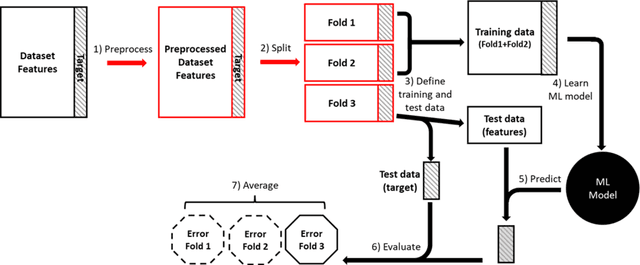
Abstract:Machine learning (ML) provides powerful tools for predictive modeling. ML's popularity stems from the promise of sample-level prediction with applications across a variety of fields from physics and marketing to healthcare. However, if not properly implemented and evaluated, ML pipelines may contain leakage typically resulting in overoptimistic performance estimates and failure to generalize to new data. This can have severe negative financial and societal implications. Our aim is to expand understanding associated with causes leading to leakage when designing, implementing, and evaluating ML pipelines. Illustrated by concrete examples, we provide a comprehensive overview and discussion of various types of leakage that may arise in ML pipelines.
Julearn: an easy-to-use library for leakage-free evaluation and inspection of ML models
Oct 19, 2023Abstract:The fast-paced development of machine learning (ML) methods coupled with its increasing adoption in research poses challenges for researchers without extensive training in ML. In neuroscience, for example, ML can help understand brain-behavior relationships, diagnose diseases, and develop biomarkers using various data sources like magnetic resonance imaging and electroencephalography. The primary objective of ML is to build models that can make accurate predictions on unseen data. Researchers aim to prove the existence of such generalizable models by evaluating performance using techniques such as cross-validation (CV), which uses systematic subsampling to estimate the generalization performance. Choosing a CV scheme and evaluating an ML pipeline can be challenging and, if used improperly, can lead to overestimated results and incorrect interpretations. We created julearn, an open-source Python library, that allow researchers to design and evaluate complex ML pipelines without encountering in common pitfalls. In this manuscript, we present the rationale behind julearn's design, its core features, and showcase three examples of previously-published research projects that can be easily implemented using this novel library. Julearn aims to simplify the entry into the ML world by providing an easy-to-use environment with built in guards against some of the most common ML pitfalls. With its design, unique features and simple interface, it poses as a useful Python-based library for research projects.
Confound-leakage: Confound Removal in Machine Learning Leads to Leakage
Oct 17, 2022Abstract:Machine learning (ML) approaches to data analysis are now widely adopted in many fields including epidemiology and medicine. To apply these approaches, confounds must first be removed as is commonly done by featurewise removal of their variance by linear regression before applying ML. Here, we show this common approach to confound removal biases ML models, leading to misleading results. Specifically, this common deconfounding approach can leak information such that what are null or moderate effects become amplified to near-perfect prediction when nonlinear ML approaches are subsequently applied. We identify and evaluate possible mechanisms for such confound-leakage and provide practical guidance to mitigate its negative impact. We demonstrate the real-world importance of confound-leakage by analyzing a clinical dataset where accuracy is overestimated for predicting attention deficit hyperactivity disorder (ADHD) with depression as a confound. Our results have wide-reaching implications for implementation and deployment of ML workflows and beg caution against na\"ive use of standard confound removal approaches.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge