Empirical Comparison between Cross-Validation and Mutation-Validation in Model Selection
Paper and Code
Nov 23, 2023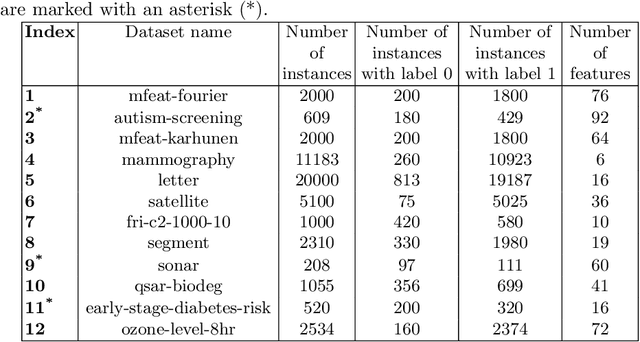
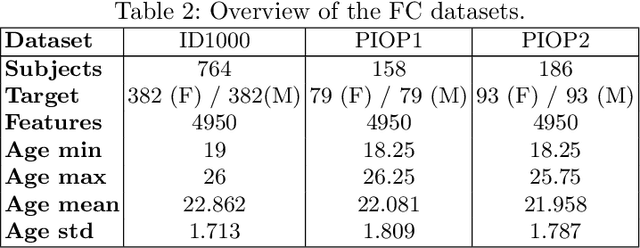
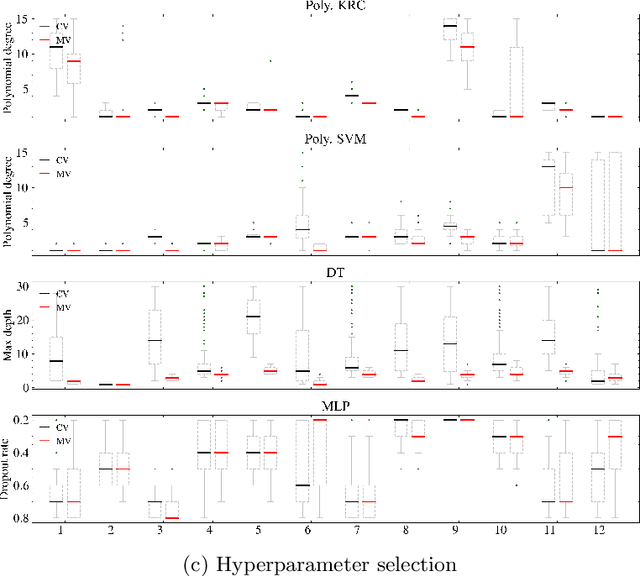
Mutation validation (MV) is a recently proposed approach for model selection, garnering significant interest due to its unique characteristics and potential benefits compared to the widely used cross-validation (CV) method. In this study, we empirically compared MV and $k$-fold CV using benchmark and real-world datasets. By employing Bayesian tests, we compared generalization estimates yielding three posterior probabilities: practical equivalence, CV superiority, and MV superiority. We also evaluated the differences in the capacity of the selected models and computational efficiency. We found that both MV and CV select models with practically equivalent generalization performance across various machine learning algorithms and the majority of benchmark datasets. MV exhibited advantages in terms of selecting simpler models and lower computational costs. However, in some cases MV selected overly simplistic models leading to underfitting and showed instability in hyperparameter selection. These limitations of MV became more evident in the evaluation of a real-world neuroscientific task of predicting sex at birth using brain functional connectivity.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge