Sambit Panda
Learning sources of variability from high-dimensional observational studies
Jul 26, 2023Abstract:Causal inference studies whether the presence of a variable influences an observed outcome. As measured by quantities such as the "average treatment effect," this paradigm is employed across numerous biological fields, from vaccine and drug development to policy interventions. Unfortunately, the majority of these methods are often limited to univariate outcomes. Our work generalizes causal estimands to outcomes with any number of dimensions or any measurable space, and formulates traditional causal estimands for nominal variables as causal discrepancy tests. We propose a simple technique for adjusting universally consistent conditional independence tests and prove that these tests are universally consistent causal discrepancy tests. Numerical experiments illustrate that our method, Causal CDcorr, leads to improvements in both finite sample validity and power when compared to existing strategies. Our methods are all open source and available at github.com/ebridge2/cdcorr.
Streaming Decision Trees and Forests
Oct 16, 2021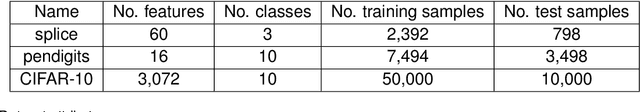
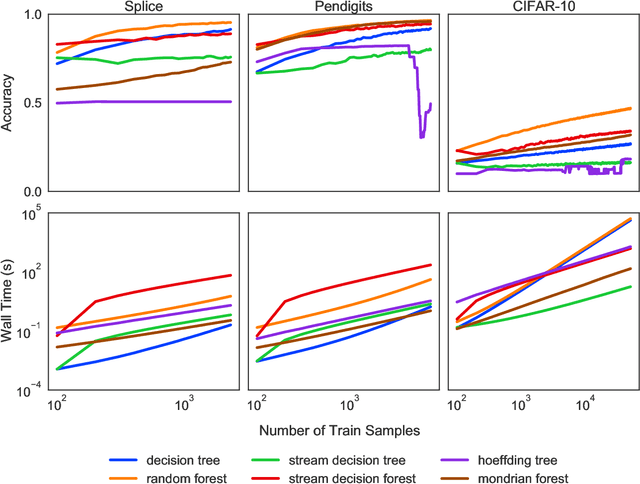
Abstract:Machine learning has successfully leveraged modern data and provided computational solutions to innumerable real-world problems, including physical and biomedical discoveries. Currently, estimators could handle both scenarios with all samples available and situations requiring continuous updates. However, there is still room for improvement on streaming algorithms based on batch decision trees and random forests, which are the leading methods in batch data tasks. In this paper, we explore the simplest partial fitting algorithm to extend batch trees and test our models: stream decision tree (SDT) and stream decision forest (SDF) on three classification tasks of varying complexities. For reference, both existing streaming trees (Hoeffding trees and Mondrian forests) and batch estimators are included in the experiments. In all three tasks, SDF consistently produces high accuracy, whereas existing estimators encounter space restraints and accuracy fluctuations. Thus, our streaming trees and forests show great potential for further improvements, which are good candidates for solving problems like distribution drift and transfer learning.
When are Deep Networks really better than Random Forests at small sample sizes?
Aug 31, 2021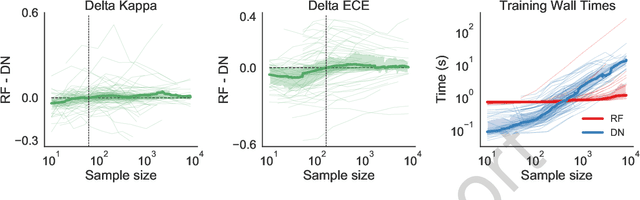

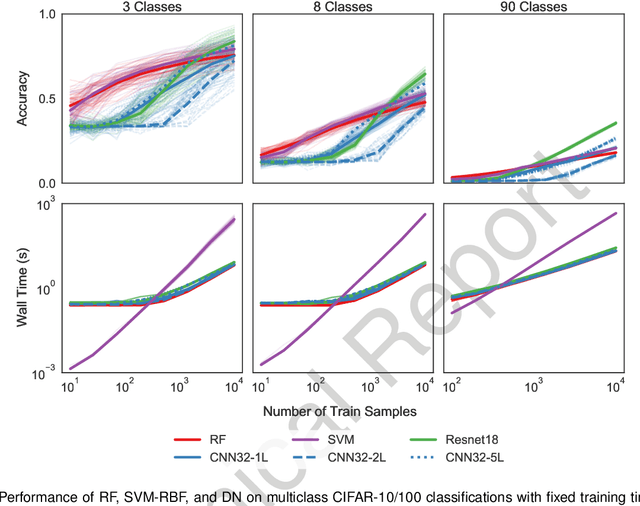
Abstract:Random forests (RF) and deep networks (DN) are two of the most popular machine learning methods in the current scientific literature and yield differing levels of performance on different data modalities. We wish to further explore and establish the conditions and domains in which each approach excels, particularly in the context of sample size and feature dimension. To address these issues, we tested the performance of these approaches across tabular, image, and audio settings using varying model parameters and architectures. Our focus is on datasets with at most 10,000 samples, which represent a large fraction of scientific and biomedical datasets. In general, we found RF to excel at tabular and structured data (image and audio) with small sample sizes, whereas DN performed better on structured data with larger sample sizes. Although we plan to continue updating this technical report in the coming months, we believe the current preliminary results may be of interest to others.
mgcpy: A Comprehensive High Dimensional Independence Testing Python Package
Jul 18, 2019
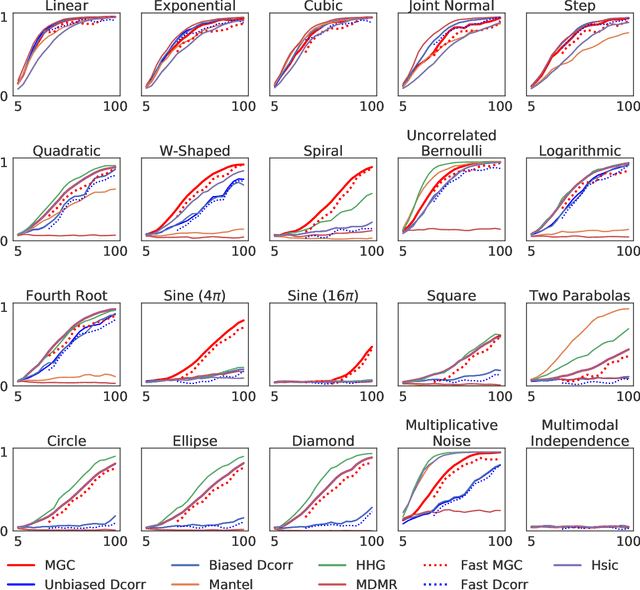
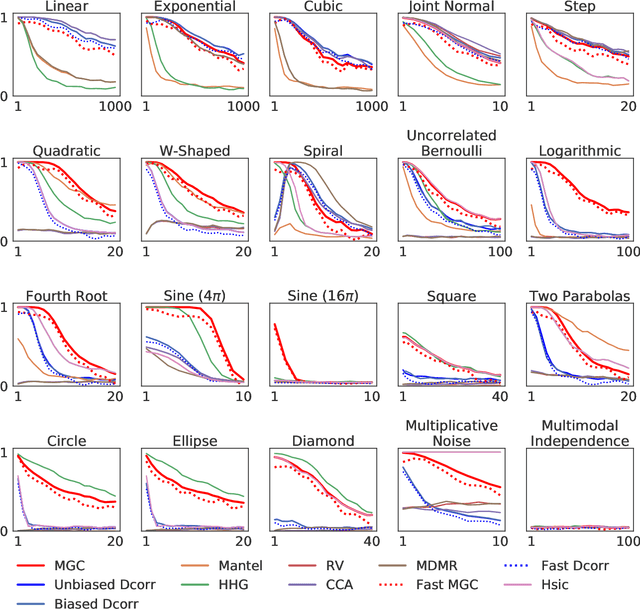
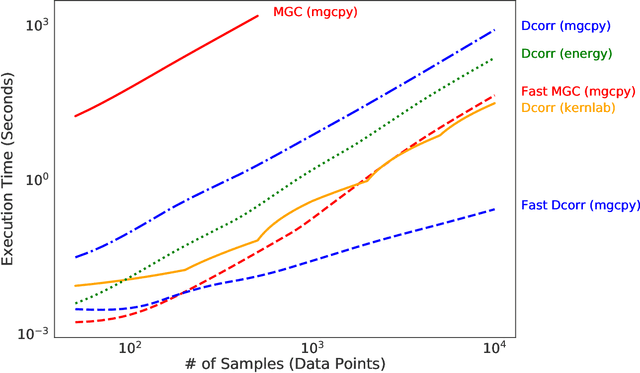
Abstract:With the increase in the amount of data in many fields, a method to consistently and efficiently decipher relationships within high dimensional data sets is important. Because many modern datasets are high-dimensional, univariate independence tests are not applicable. While many multivariate independence tests have R packages available, the interfaces are inconsistent, most are not available in Python. mgcpy is an extensive Python library that includes many state of the art high-dimensional independence testing procedures using a common interface. The package is easy-to-use and is flexible enough to enable future extensions. This manuscript provides details for each of the tests as well as extensive power and run-time benchmarks on a suite of high-dimensional simulations previously used in different publications. The appendix includes demonstrations of how the user can interact with the package, as well as links and documentation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge