Sahil Manocha
SketchParse : Towards Rich Descriptions for Poorly Drawn Sketches using Multi-Task Hierarchical Deep Networks
Sep 05, 2017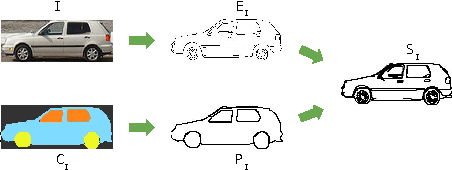

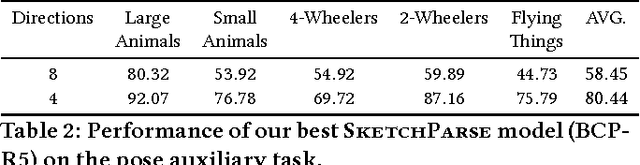
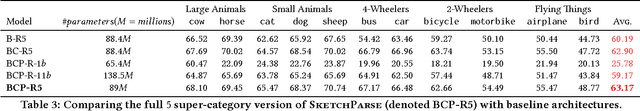
Abstract:The ability to semantically interpret hand-drawn line sketches, although very challenging, can pave way for novel applications in multimedia. We propose SketchParse, the first deep-network architecture for fully automatic parsing of freehand object sketches. SketchParse is configured as a two-level fully convolutional network. The first level contains shared layers common to all object categories. The second level contains a number of expert sub-networks. Each expert specializes in parsing sketches from object categories which contain structurally similar parts. Effectively, the two-level configuration enables our architecture to scale up efficiently as additional categories are added. We introduce a router layer which (i) relays sketch features from shared layers to the correct expert (ii) eliminates the need to manually specify object category during inference. To bypass laborious part-level annotation, we sketchify photos from semantic object-part image datasets and use them for training. Our architecture also incorporates object pose prediction as a novel auxiliary task which boosts overall performance while providing supplementary information regarding the sketch. We demonstrate SketchParse's abilities (i) on two challenging large-scale sketch datasets (ii) in parsing unseen, semantically related object categories (iii) in improving fine-grained sketch-based image retrieval. As a novel application, we also outline how SketchParse's output can be used to generate caption-style descriptions for hand-drawn sketches.
Enhancing the functional content of protein interaction networks
Oct 25, 2012



Abstract:Protein interaction networks are a promising type of data for studying complex biological systems. However, despite the rich information embedded in these networks, they face important data quality challenges of noise and incompleteness that adversely affect the results obtained from their analysis. Here, we explore the use of the concept of common neighborhood similarity (CNS), which is a form of local structure in networks, to address these issues. Although several CNS measures have been proposed in the literature, an understanding of their relative efficacies for the analysis of interaction networks has been lacking. We follow the framework of graph transformation to convert the given interaction network into a transformed network corresponding to a variety of CNS measures evaluated. The effectiveness of each measure is then estimated by comparing the quality of protein function predictions obtained from its corresponding transformed network with those from the original network. Using a large set of S. cerevisiae interactions, and a set of 136 GO terms, we find that several of the transformed networks produce more accurate predictions than those obtained from the original network. In particular, the $HC.cont$ measure proposed here performs particularly well for this task. Further investigation reveals that the two major factors contributing to this improvement are the abilities of CNS measures, especially $HC.cont$, to prune out noisy edges and introduce new links between functionally related proteins.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge