Runpu Wei
OCORD: Open-Campus Object Removal Dataset
Jan 13, 2025Abstract:The rapid advancements in generative models, particularly diffusion-based techniques, have revolutionized image inpainting tasks by enabling the generation of high-fidelity and diverse content. However, object removal remains under-explored as a specific subset of inpainting, facing challenges such as inadequate semantic understanding and the unintended generation of artifacts. Existing datasets for object removal often rely on synthetic data, which fails to align with real-world scenarios, limiting model performance. Although some real-world datasets address these issues partially, they suffer from scalability, annotation inefficiencies, and limited realism in physical phenomena such as lighting and shadows. To address these limitations, this paper introduces a novel approach to object removal by constructing a high-resolution real-world dataset through long-duration video capture with fixed camera settings. Leveraging advanced tools such as Grounding-DINO, Segment-Anything-Model, and MASA for automated annotation, we provides image, background, and mask pairs while significantly reducing annotation time and labor. With our efficient annotation pipeline, we release the first fully open, high-resolution real-world dataset for object removal, and improved performance in object removal tasks through fine-tuning of pre-trained diffusion models.
Polyp-E: Benchmarking the Robustness of Deep Segmentation Models via Polyp Editing
Oct 22, 2024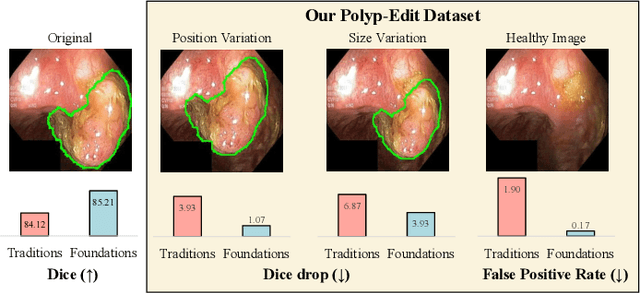
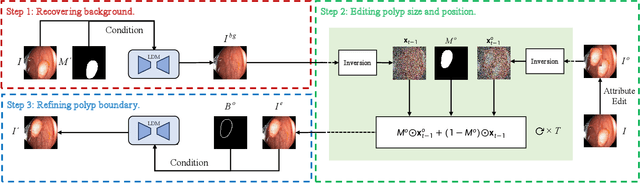
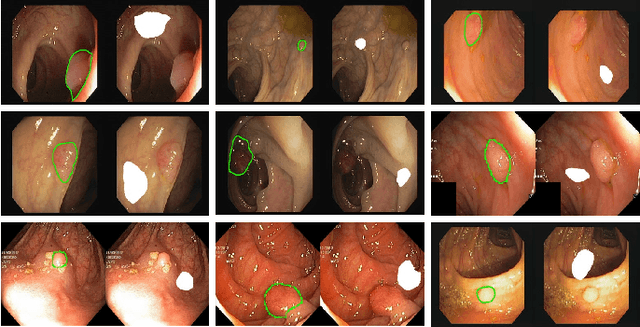
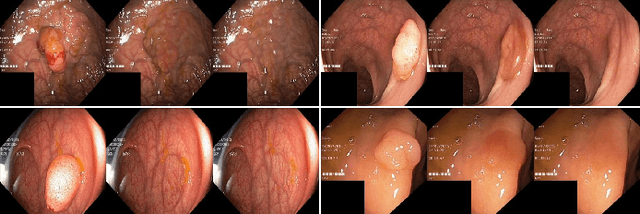
Abstract:Automatic polyp segmentation is helpful to assist clinical diagnosis and treatment. In daily clinical practice, clinicians exhibit robustness in identifying polyps with both location and size variations. It is uncertain if deep segmentation models can achieve comparable robustness in automated colonoscopic analysis. To benchmark the model robustness, we focus on evaluating the robustness of segmentation models on the polyps with various attributes (e.g. location and size) and healthy samples. Based on the Latent Diffusion Model, we perform attribute editing on real polyps and build a new dataset named Polyp-E. Our synthetic dataset boasts exceptional realism, to the extent that clinical experts find it challenging to discern them from real data. We evaluate several existing polyp segmentation models on the proposed benchmark. The results reveal most of the models are highly sensitive to attribute variations. As a novel data augmentation technique, the proposed editing pipeline can improve both in-distribution and out-of-distribution generalization ability. The code and datasets will be released.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge