Ruheng Wang
Scaling Medical Reasoning Verification via Tool-Integrated Reinforcement Learning
Jan 28, 2026Abstract:Large language models have achieved strong performance on medical reasoning benchmarks, yet their deployment in clinical settings demands rigorous verification to ensure factual accuracy. While reward models offer a scalable approach for reasoning trace verification, existing methods face two limitations: they produce only scalar reward values without explicit justification, and they rely on single-pass retrieval that precludes adaptive knowledge access as verification unfolds. We introduce $\method$, an agentic framework that addresses these limitations by training medical reasoning verifiers to iteratively query external medical corpora during evaluation. Our approach combines tool-augmented verification with an iterative reinforcement learning paradigm that requires only trace-level supervision, alongside an adaptive curriculum mechanism that dynamically adjusts training data distribution. Across four medical reasoning benchmarks, $\method$ achieves substantial gains over existing methods, improving MedQA accuracy by 23.5% and MedXpertQA by 32.0% relative to the base generator in particular. Crucially, $\method$ demonstrates an $\mathbf{8\times}$ reduction in sampling budget requirement compared to prior reward model baselines. These findings establish that grounding verification in dynamically retrieved evidence offers a principled path toward more reliable medical reasoning systems.
PepThink-R1: LLM for Interpretable Cyclic Peptide Optimization with CoT SFT and Reinforcement Learning
Aug 20, 2025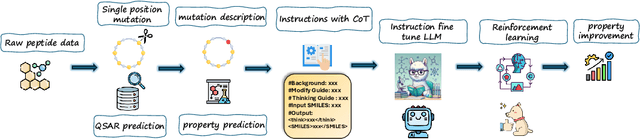



Abstract:Designing therapeutic peptides with tailored properties is hindered by the vastness of sequence space, limited experimental data, and poor interpretability of current generative models. To address these challenges, we introduce PepThink-R1, a generative framework that integrates large language models (LLMs) with chain-of-thought (CoT) supervised fine-tuning and reinforcement learning (RL). Unlike prior approaches, PepThink-R1 explicitly reasons about monomer-level modifications during sequence generation, enabling interpretable design choices while optimizing for multiple pharmacological properties. Guided by a tailored reward function balancing chemical validity and property improvements, the model autonomously explores diverse sequence variants. We demonstrate that PepThink-R1 generates cyclic peptides with significantly enhanced lipophilicity, stability, and exposure, outperforming existing general LLMs (e.g., GPT-5) and domain-specific baseline in both optimization success and interpretability. To our knowledge, this is the first LLM-based peptide design framework that combines explicit reasoning with RL-driven property control, marking a step toward reliable and transparent peptide optimization for therapeutic discovery.
Multi-view deep learning based molecule design and structural optimization accelerates the SARS-CoV-2 inhibitor discovery
Dec 03, 2022Abstract:In this work, we propose MEDICO, a Multi-viEw Deep generative model for molecule generation, structural optimization, and the SARS-CoV-2 Inhibitor disCOvery. To the best of our knowledge, MEDICO is the first-of-this-kind graph generative model that can generate molecular graphs similar to the structure of targeted molecules, with a multi-view representation learning framework to sufficiently and adaptively learn comprehensive structural semantics from targeted molecular topology and geometry. We show that our MEDICO significantly outperforms the state-of-the-art methods in generating valid, unique, and novel molecules under benchmarking comparisons. In particular, we showcase the multi-view deep learning model enables us to generate not only the molecules structurally similar to the targeted molecules but also the molecules with desired chemical properties, demonstrating the strong capability of our model in exploring the chemical space deeply. Moreover, case study results on targeted molecule generation for the SARS-CoV-2 main protease (Mpro) show that by integrating molecule docking into our model as chemical priori, we successfully generate new small molecules with desired drug-like properties for the Mpro, potentially accelerating the de novo design of Covid-19 drugs. Further, we apply MEDICO to the structural optimization of three well-known Mpro inhibitors (N3, 11a, and GC376) and achieve ~88% improvement in their binding affinity to Mpro, demonstrating the application value of our model for the development of therapeutics for SARS-CoV-2 infection.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge