Roberto Novoa
SkinCon: A skin disease dataset densely annotated by domain experts for fine-grained model debugging and analysis
Feb 01, 2023



Abstract:For the deployment of artificial intelligence (AI) in high-risk settings, such as healthcare, methods that provide interpretability/explainability or allow fine-grained error analysis are critical. Many recent methods for interpretability/explainability and fine-grained error analysis use concepts, which are meta-labels that are semantically meaningful to humans. However, there are only a few datasets that include concept-level meta-labels and most of these meta-labels are relevant for natural images that do not require domain expertise. Densely annotated datasets in medicine focused on meta-labels that are relevant to a single disease such as melanoma. In dermatology, skin disease is described using an established clinical lexicon that allows clinicians to describe physical exam findings to one another. To provide a medical dataset densely annotated by domain experts with annotations useful across multiple disease processes, we developed SkinCon: a skin disease dataset densely annotated by dermatologists. SkinCon includes 3230 images from the Fitzpatrick 17k dataset densely annotated with 48 clinical concepts, 22 of which have at least 50 images representing the concept. The concepts used were chosen by two dermatologists considering the clinical descriptor terms used to describe skin lesions. Examples include "plaque", "scale", and "erosion". The same concepts were also used to label 656 skin disease images from the Diverse Dermatology Images dataset, providing an additional external dataset with diverse skin tone representations. We review the potential applications for the SkinCon dataset, such as probing models, concept-based explanations, and concept bottlenecks. Furthermore, we use SkinCon to demonstrate two of these use cases: debugging mistakes of an existing dermatology AI model with concepts and developing interpretable models with post-hoc concept bottleneck models.
Development and Clinical Evaluation of an AI Support Tool for Improving Telemedicine Photo Quality
Sep 12, 2022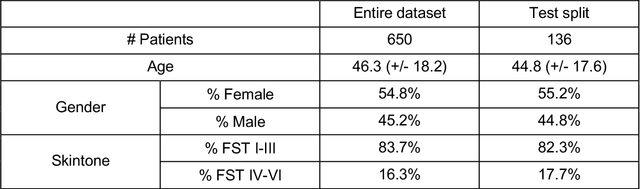
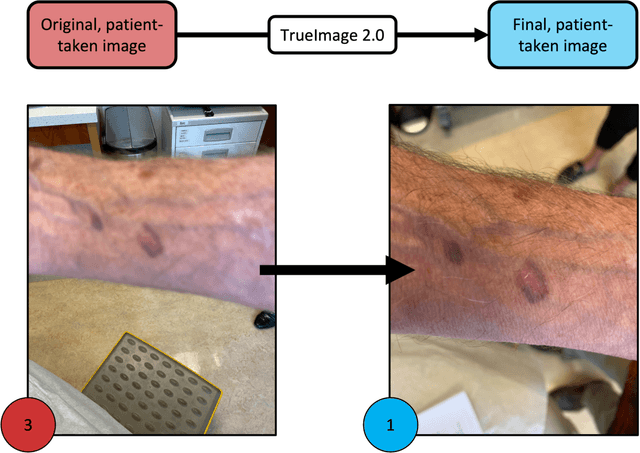
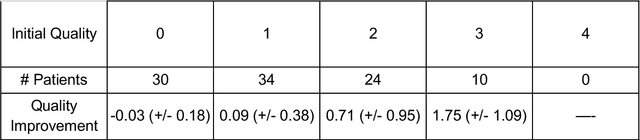
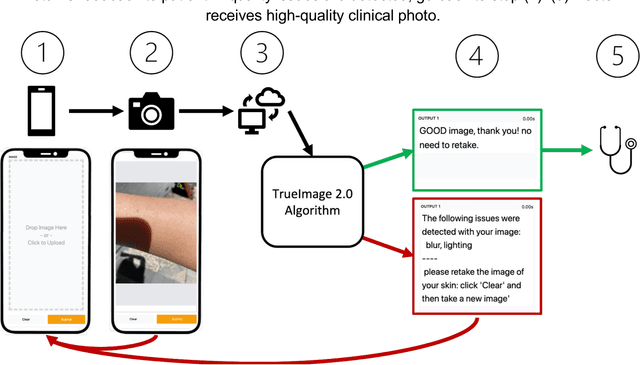
Abstract:Telemedicine utilization was accelerated during the COVID-19 pandemic, and skin conditions were a common use case. However, the quality of photographs sent by patients remains a major limitation. To address this issue, we developed TrueImage 2.0, an artificial intelligence (AI) model for assessing patient photo quality for telemedicine and providing real-time feedback to patients for photo quality improvement. TrueImage 2.0 was trained on 1700 telemedicine images annotated by clinicians for photo quality. On a retrospective dataset of 357 telemedicine images, TrueImage 2.0 effectively identified poor quality images (Receiver operator curve area under the curve (ROC-AUC) =0.78) and the reason for poor quality (Blurry ROC-AUC=0.84, Lighting issues ROC-AUC=0.70). The performance is consistent across age, gender, and skin tone. Next, we assessed whether patient-TrueImage 2.0 interaction led to an improvement in submitted photo quality through a prospective clinical pilot study with 98 patients. TrueImage 2.0 reduced the number of patients with a poor-quality image by 68.0%.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge