Rita Brugarolas Brufau
BatchGNN: Efficient CPU-Based Distributed GNN Training on Very Large Graphs
Jun 23, 2023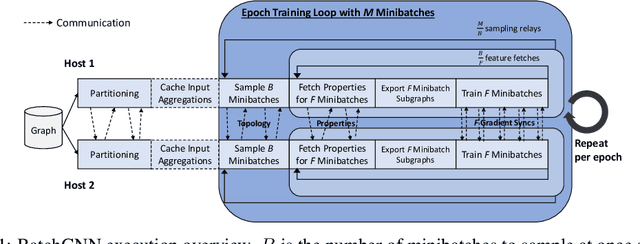



Abstract:We present BatchGNN, a distributed CPU system that showcases techniques that can be used to efficiently train GNNs on terabyte-sized graphs. It reduces communication overhead with macrobatching in which multiple minibatches' subgraph sampling and feature fetching are batched into one communication relay to reduce redundant feature fetches when input features are static. BatchGNN provides integrated graph partitioning and native GNN layer implementations to improve runtime, and it can cache aggregated input features to further reduce sampling overhead. BatchGNN achieves an average $3\times$ speedup over DistDGL on three GNN models trained on OGBN graphs, outperforms the runtimes reported by distributed GPU systems $P^3$ and DistDGLv2, and scales to a terabyte-sized graph.
Learning Reduced-Order Models for Cardiovascular Simulations with Graph Neural Networks
Mar 13, 2023



Abstract:Reduced-order models based on physics are a popular choice in cardiovascular modeling due to their efficiency, but they may experience reduced accuracy when working with anatomies that contain numerous junctions or pathological conditions. We develop one-dimensional reduced-order models that simulate blood flow dynamics using a graph neural network trained on three-dimensional hemodynamic simulation data. Given the initial condition of the system, the network iteratively predicts the pressure and flow rate at the vessel centerline nodes. Our numerical results demonstrate the accuracy and generalizability of our method in physiological geometries comprising a variety of anatomies and boundary conditions. Our findings demonstrate that our approach can achieve errors below 2% and 3% for pressure and flow rate, respectively, provided there is adequate training data. As a result, our method exhibits superior performance compared to physics-based one-dimensional models, while maintaining high efficiency at inference time.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge