Rise Ooi
Controllability, Multiplexing, and Transfer Learning in Networks using Evolutionary Learning
Nov 14, 2018



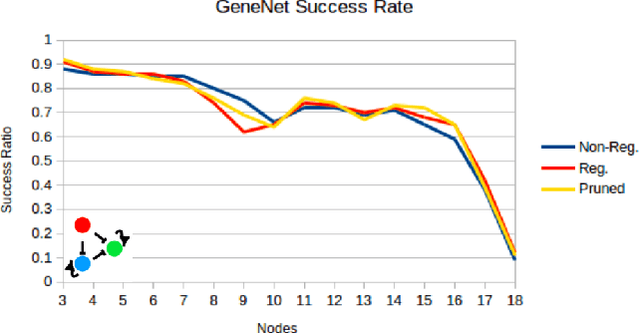
Abstract:Networks are fundamental building blocks for representing data, and computations. Remarkable progress in learning in structurally defined (shallow or deep) networks has recently been achieved. Here we introduce evolutionary exploratory search and learning method of topologically flexible networks under the constraint of producing elementary computational steady-state input-output operations. Our results include; (1) the identification of networks, over four orders of magnitude, implementing computation of steady-state input-output functions, such as a band-pass filter, a threshold function, and an inverse band-pass function. Next, (2) the learned networks are technically controllable as only a small number of driver nodes are required to move the system to a new state. Furthermore, we find that the fraction of required driver nodes is constant during evolutionary learning, suggesting a stable system design. (3), our framework allows multiplexing of different computations using the same network. For example, using a binary representation of the inputs, the network can readily compute three different input-output functions. Finally, (4) the proposed evolutionary learning demonstrates transfer learning. If the system learns one function A, then learning B requires on average less number of steps as compared to learning B from tabula rasa. We conclude that the constrained evolutionary learning produces large robust controllable circuits, capable of multiplexing and transfer learning. Our study suggests that network-based computations of steady-state functions, representing either cellular modules of cell-to-cell communication networks or internal molecular circuits communicating within a cell, could be a powerful model for biologically inspired computing. This complements conceptualizations such as attractor based models, or reservoir computing.
Learning Functions in Large Networks requires Modularity and produces Multi-Agent Dynamics
Aug 21, 2018
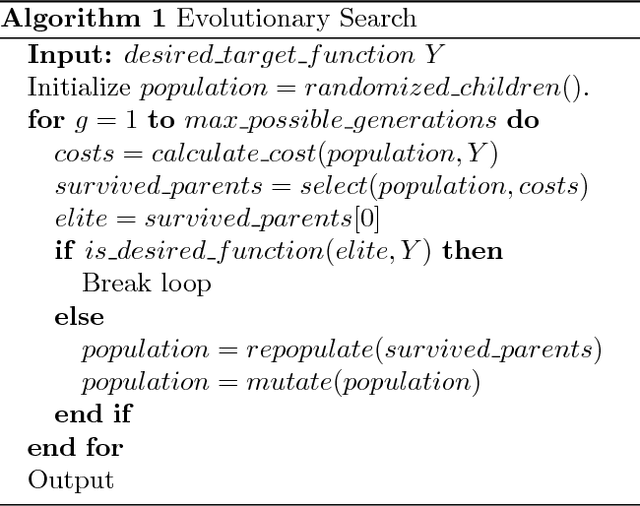

Abstract:Networks are abundant in biological systems. Small sized over-represented network motifs have been discovered, and it has been suggested that these constitute functional building blocks. We ask whether larger dynamical network motifs exist in biological networks, thus contributing to the higher-order organization of a network. To end this, we introduce a gradient descent machine learning (ML) approach and genetic algorithms to learn larger functional motifs in contrast to an (unfeasible) exhaustive search. We use the French Flag (FF) and Switch functional motif as case studies motivated from biology. While our algorithm successfully learns large functional motifs, we identify a threshold size of approximately 20 nodes beyond which learning breaks down. Therefore we investigate the stability of the motifs. We find that the size of the real negative eigenvalues of the Jacobian decreases with increasing system size, thus conferring instability. Finally, without imposing learning an input-output for all the components of the network, we observe that unconstrained middle components of the network still learn the desired function, a form of homogeneous team learning. We conclude that the size limitation of learnability, most likely due to stability constraints, impose a definite requirement for modularity in networked systems while enabling team learning within unconstrained parts of the module. Thus, the observation that community structures and modularity are abundant in biological networks could be accounted for by a computational compositional network structure.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge