Richard J. Preen
A hierarchical approach for assessing the vulnerability of tree-based classification models to membership inference attack
Feb 13, 2025Abstract:Machine learning models can inadvertently expose confidential properties of their training data, making them vulnerable to membership inference attacks (MIA). While numerous evaluation methods exist, many require computationally expensive processes, such as training multiple shadow models. This article presents two new complementary approaches for efficiently identifying vulnerable tree-based models: an ante-hoc analysis of hyperparameter choices and a post-hoc examination of trained model structure. While these new methods cannot certify whether a model is safe from MIA, they provide practitioners with a means to significantly reduce the number of models that need to undergo expensive MIA assessment through a hierarchical filtering approach. More specifically, it is shown that the rank order of disclosure risk for different hyperparameter combinations remains consistent across datasets, enabling the development of simple, human-interpretable rules for identifying relatively high-risk models before training. While this ante-hoc analysis cannot determine absolute safety since this also depends on the specific dataset, it allows the elimination of unnecessarily risky configurations during hyperparameter tuning. Additionally, computationally inexpensive structural metrics serve as indicators of MIA vulnerability, providing a second filtering stage to identify risky models after training but before conducting expensive attacks. Empirical results show that hyperparameter-based risk prediction rules can achieve high accuracy in predicting the most at risk combinations of hyperparameters across different tree-based model types, while requiring no model training. Moreover, target model accuracy is not seen to correlate with privacy risk, suggesting opportunities to optimise model configurations for both performance and privacy.
Safe machine learning model release from Trusted Research Environments: The AI-SDC package
Dec 06, 2022Abstract:We present AI-SDC, an integrated suite of open source Python tools to facilitate Statistical Disclosure Control (SDC) of Machine Learning (ML) models trained on confidential data prior to public release. AI-SDC combines (i) a SafeModel package that extends commonly used ML models to provide ante-hoc SDC by assessing the vulnerability of disclosure posed by the training regime; and (ii) an Attacks package that provides post-hoc SDC by rigorously assessing the empirical disclosure risk of a model through a variety of simulated attacks after training. The AI-SDC code and documentation are available under an MIT license at https://github.com/AI-SDC/AI-SDC.
ACRO: A multi-language toolkit for supporting Automated Checking of Research Outputs
Dec 06, 2022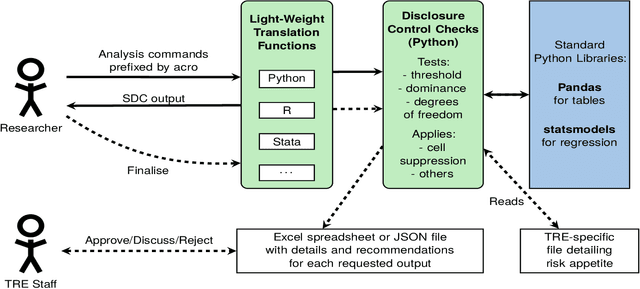
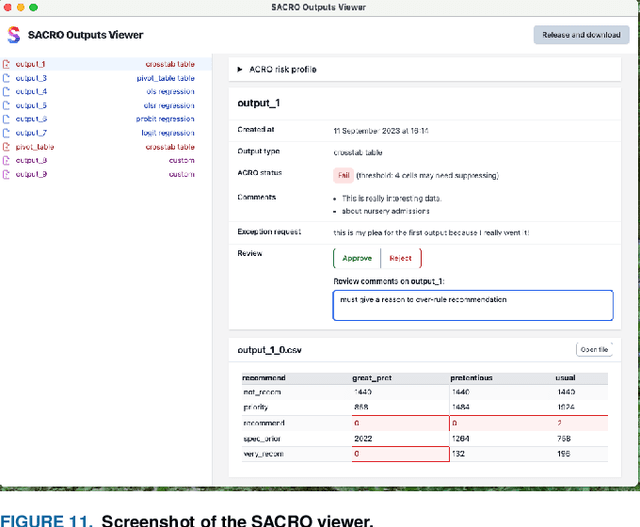
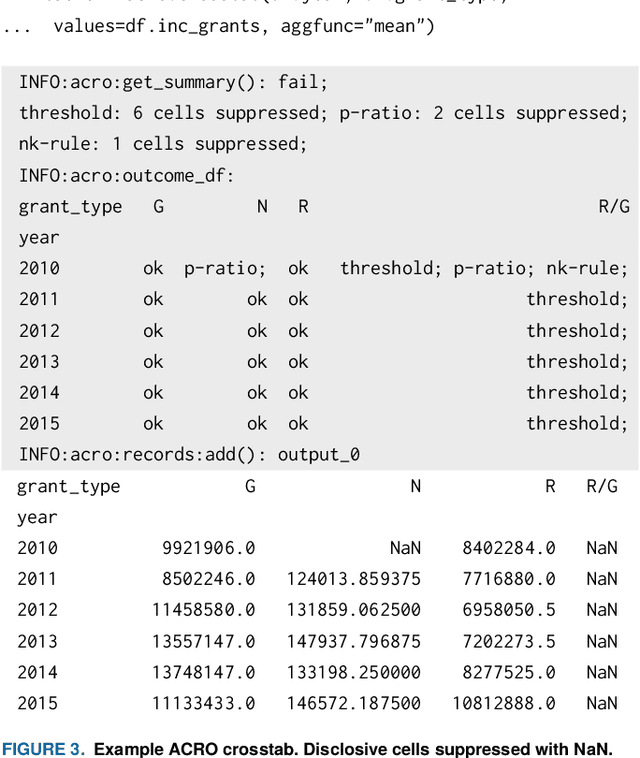
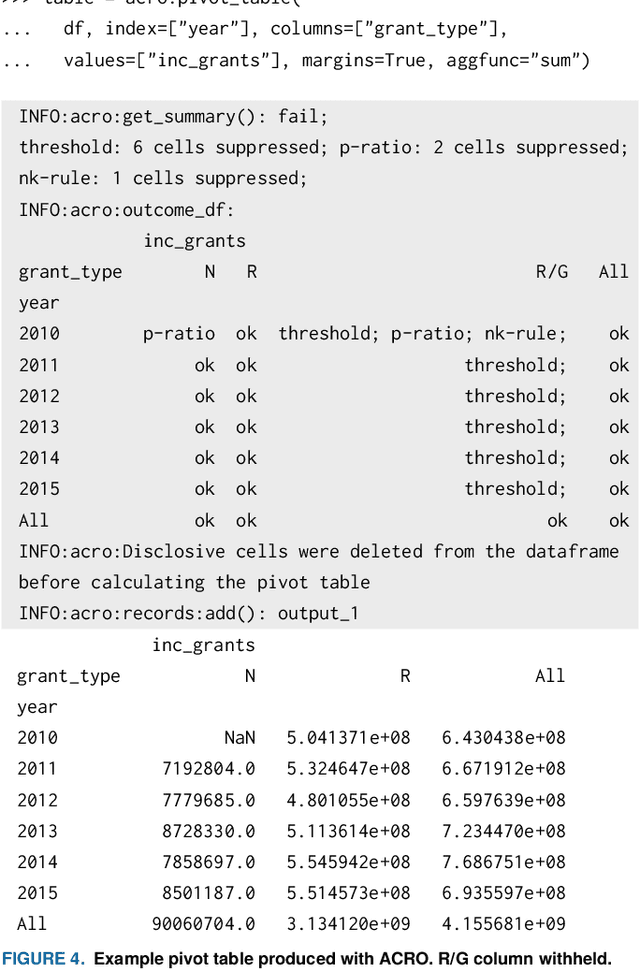
Abstract:This paper discusses the development of an open source tool ACRO, (Automatic Checking of Research Outputs) to assist researchers and data governance teams by distinguishing between: research output that is safe to publish; output that requires further analysis; and output that cannot be published because it creates substantial risk of disclosing private data. ACRO extends the functionality and accessibility of a previous prototype by providing a light-weight 'skin' that sits over well-known analysis tools, and enables access in a variety of programming languages researchers might use. This adds functionality to (i) identify potentially disclosive outputs against a range of commonly used disclosure tests; (ii) suppress outputs where required; (iii) report reasons for suppression; and (iv) produce simple summary documents Trusted Research Environment (TRE) staff can use to streamline their workflow. The ACRO code and documentation are available under an MIT license at https://github.com/AI-SDC/ACRO
Deep Learning with a Classifier System: Initial Results
Mar 01, 2021


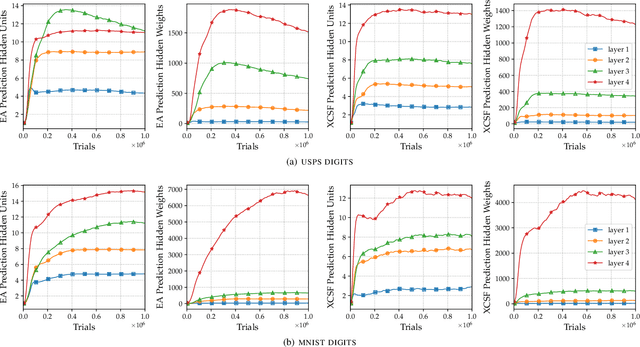
Abstract:This article presents the first results from using a learning classifier system capable of performing adaptive computation with deep neural networks. Individual classifiers within the population are composed of two neural networks. The first acts as a gating or guarding component, which enables the conditional computation of an associated deep neural network on a per instance basis. Self-adaptive mutation is applied upon reproduction and prediction networks are refined with stochastic gradient descent during lifetime learning. The use of fully-connected and convolutional layers are evaluated on handwritten digit recognition tasks where evolution adapts (i) the gradient descent learning rate applied to each layer (ii) the number of units within each layer, i.e., the number of fully-connected neurons and the number of convolutional kernel filters (iii) the connectivity of each layer, i.e., whether each weight is active (iv) the weight magnitudes, enabling escape from local optima. The system automatically reduces the number of weights and units while maintaining performance after achieving a maximum prediction error.
Autoencoding with XCSF
Nov 05, 2019



Abstract:Autoencoders enable data dimensionality reduction and are a key component of many (deep) learning systems. This article explores the use of the XCSF online evolutionary reinforcement learning system to perform autoencoding. Initial results using a neural network representation and combining artificial evolution with stochastic gradient descent, suggest it is an effective approach to data reduction. The approach adaptively subdivides the input domain into local approximations that are simpler than a global neural network solution. By allowing the number of neurons in the autoencoders to evolve, this further enables the emergence of an ensemble of structurally heterogeneous solutions to cover the problem space. In this case, networks of differing complexity are typically seen to cover different areas of the problem space. Furthermore, the rate of gradient descent applied to each layer is tuned via self-adaptive mutation, thereby reducing the parameter optimisation task.
Towards an Evolvable Cancer Treatment Simulator
Dec 19, 2018



Abstract:The use of high-fidelity computational simulations promises to enable high-throughput hypothesis testing and optimisation of cancer therapies. However, increasing realism comes at the cost of increasing computational requirements. This article explores the use of surrogate-assisted evolutionary algorithms to optimise the targeted delivery of a therapeutic compound to cancerous tumour cells with the multicellular simulator, PhysiCell. The use of both Gaussian process models and multi-layer perceptron neural network surrogate models are investigated. We find that evolutionary algorithms are able to effectively explore the parameter space of biophysical properties within the agent-based simulations, minimising the resulting number of cancerous cells after a period of simulated treatment. Both model-assisted algorithms are found to outperform a standard evolutionary algorithm, demonstrating their ability to perform a more effective search within the very small evaluation budget.
Evolutionary n-level Hypergraph Partitioning with Adaptive Coarsening
Oct 08, 2018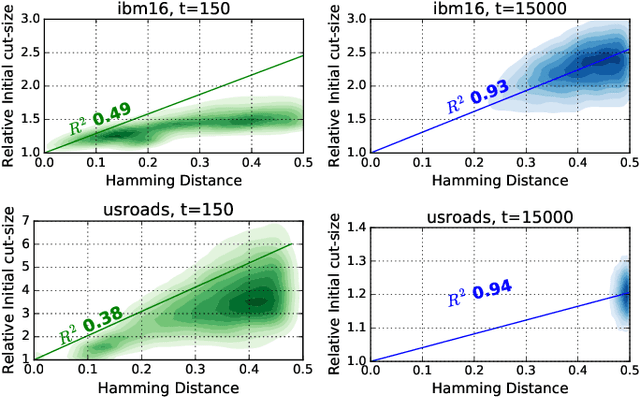
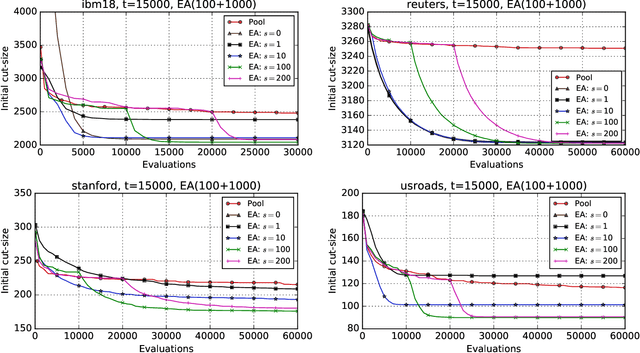
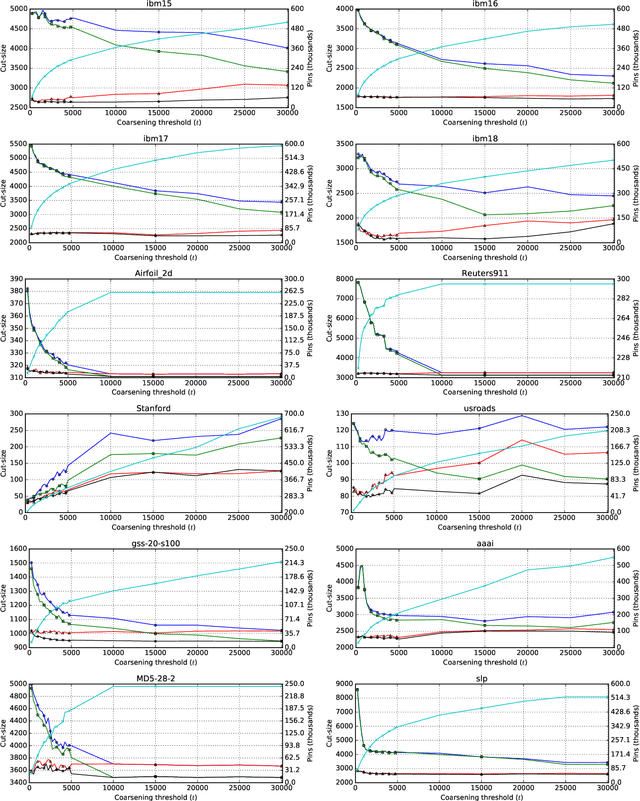
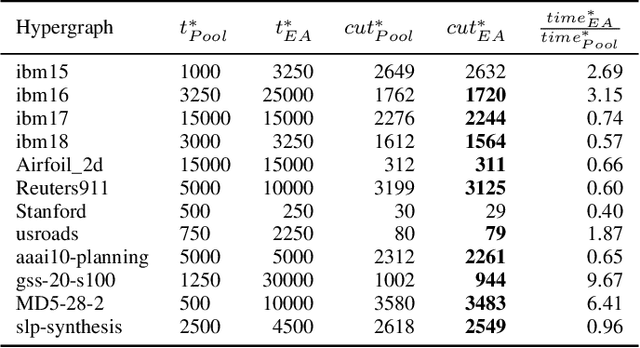
Abstract:Hypergraph partitioning is an NP-hard problem that occurs in many computer science applications where it is necessary to reduce large problems into a number of smaller, computationally tractable sub-problems. Current techniques use a multilevel approach wherein an initial partitioning is performed after compressing the hypergraph to a predetermined level. This level is typically chosen to produce very coarse hypergraphs in which heuristic algorithms are fast and effective. This article presents a novel memetic algorithm which remains effective on larger initial hypergraphs. This enables the exploitation of information that can be lost during coarsening and results in improved final solution quality. We use this algorithm to present an empirical analysis of the space of possible initial hypergraphs in terms of its searchability at different levels of coarsening. We find that the best results arise at coarsening levels unique to each hypergraph. Based on this, we introduce an adaptive scheme that stops coarsening when the rate of information loss in a hypergraph becomes non-linear and show that this produces further improvements. The results show that we have identified a valuable role for evolutionary algorithms within the current state-of-the-art hypergraph partitioning framework.
Design Mining Microbial Fuel Cell Cascades
Jan 31, 2017



Abstract:Microbial fuel cells (MFCs) perform wastewater treatment and electricity production through the conversion of organic matter using microorganisms. For practical applications, it has been suggested that greater efficiency can be achieved by arranging multiple MFC units into physical stacks in a cascade with feedstock flowing sequentially between units. In this paper, we investigate the use of computational intelligence to physically explore and optimise (potentially) heterogeneous MFC designs in a cascade, i.e. without simulation. Conductive structures are 3-D printed and inserted into the anodic chamber of each MFC unit, augmenting a carbon fibre veil anode and affecting the hydrodynamics, including the feedstock volume and hydraulic retention time, as well as providing unique habitats for microbial colonisation. We show that it is possible to use design mining to identify new conductive inserts that increase both the cascade power output and power density.
On Design Mining: Coevolution and Surrogate Models
Nov 23, 2016Abstract:Design mining is the use of computational intelligence techniques to iteratively search and model the attribute space of physical objects evaluated directly through rapid prototyping to meet given objectives. It enables the exploitation of novel materials and processes without formal models or complex simulation. In this paper, we focus upon the coevolutionary nature of the design process when it is decomposed into concurrent sub-design threads due to the overall complexity of the task. Using an abstract, tuneable model of coevolution we consider strategies to sample sub-thread designs for whole system testing and how best to construct and use surrogate models within the coevolutionary scenario. Drawing on our findings, the paper then describes the effective design of an array of six heterogeneous vertical-axis wind turbines.
Towards the Evolution of Vertical-Axis Wind Turbines using Supershapes
Feb 15, 2016



Abstract:We have recently presented an initial study of evolutionary algorithms used to design vertical-axis wind turbines (VAWTs) wherein candidate prototypes are evaluated under approximated wind tunnel conditions after being physically instantiated by a 3D printer. That is, unlike other approaches such as computational fluid dynamics simulations, no mathematical formulations are used and no model assumptions are made. However, the representation used significantly restricted the range of morphologies explored. In this paper, we present initial explorations into the use of a simple generative encoding, known as Gielis superformula, that produces a highly flexible 3D shape representation to design VAWT. First, the target-based evolution of 3D artefacts is investigated and subsequently initial design experiments are performed wherein each VAWT candidate is physically instantiated and evaluated under approximated wind tunnel conditions. It is shown possible to produce very closely matching designs of a number of 3D objects through the evolution of supershapes produced by Gielis superformula. Moreover, it is shown possible to use artificial physical evolution to identify novel and increasingly efficient supershape VAWT designs.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge