Reinhard Furrer
Time-series Random Process Complexity Ranking Using a Bound on Conditional Differential Entropy
Oct 23, 2025Abstract:Conditional differential entropy provides an intuitive measure for relatively ranking time-series complexity by quantifying uncertainty in future observations given past context. However, its direct computation for high-dimensional processes from unknown distributions is often intractable. This paper builds on the information theoretic prediction error bounds established by Fang et al. \cite{fang2019generic}, which demonstrate that the conditional differential entropy \textbf{$h(X_k \mid X_{k-1},...,X_{k-m})$} is upper bounded by a function of the determinant of the covariance matrix of next-step prediction errors for any next step prediction model. We add to this theoretical framework by further increasing this bound by leveraging Hadamard's inequality and the positive semi-definite property of covariance matrices. To see if these bounds can be used to rank the complexity of time series, we conducted two synthetic experiments: (1) controlled linear autoregressive processes with additive Gaussian noise, where we compare ordinary least squares prediction error entropy proxies to the true entropies of various additive noises, and (2) a complexity ranking task of bio-inspired synthetic audio data with unknown entropy, where neural network prediction errors are used to recover the known complexity ordering. This framework provides a computationally tractable method for time-series complexity ranking using prediction errors from next-step prediction models, that maintains a theoretical foundation in information theory.
A class of modular and flexible covariate-based covariance functions for nonstationary spatial modeling
Oct 22, 2024Abstract:The assumptions of stationarity and isotropy often stated over spatial processes have not aged well during the last two decades, partly explained by the combination of computational developments and the increasing availability of high-resolution spatial data. While a plethora of approaches have been developed to relax these assumptions, it is often a costly tradeoff between flexibility and a diversity of computational challenges. In this paper, we present a class of covariance functions that relies on fixed, observable spatial information that provides a convenient tradeoff while offering an extra layer of numerical and visual representation of the flexible spatial dependencies. This model allows for separate parametric structures for different sources of nonstationarity, such as marginal standard deviation, geometric anisotropy, and smoothness. It simplifies to a Mat\'ern covariance function in its basic form and is adaptable for large datasets, enhancing flexibility and computational efficiency. We analyze the capabilities of the presented model through simulation studies and an application to Swiss precipitation data.
Iterative Methods for Full-Scale Gaussian Process Approximations for Large Spatial Data
May 23, 2024



Abstract:Gaussian processes are flexible probabilistic regression models which are widely used in statistics and machine learning. However, a drawback is their limited scalability to large data sets. To alleviate this, we consider full-scale approximations (FSAs) that combine predictive process methods and covariance tapering, thus approximating both global and local structures. We show how iterative methods can be used to reduce the computational costs for calculating likelihoods, gradients, and predictive distributions with FSAs. We introduce a novel preconditioner and show that it accelerates the conjugate gradient method's convergence speed and mitigates its sensitivity with respect to the FSA parameters and the eigenvalue structure of the original covariance matrix, and we demonstrate empirically that it outperforms a state-of-the-art pivoted Cholesky preconditioner. Further, we present a novel, accurate, and fast way to calculate predictive variances relying on stochastic estimations and iterative methods. In both simulated and real-world data experiments, we find that our proposed methodology achieves the same accuracy as Cholesky-based computations with a substantial reduction in computational time. Finally, we also compare different approaches for determining inducing points in predictive process and FSA models. All methods are implemented in a free C++ software library with high-level Python and R packages.
BiasBed -- Rigorous Texture Bias Evaluation
Dec 01, 2022



Abstract:The well-documented presence of texture bias in modern convolutional neural networks has led to a plethora of algorithms that promote an emphasis on shape cues, often to support generalization to new domains. Yet, common datasets, benchmarks and general model selection strategies are missing, and there is no agreed, rigorous evaluation protocol. In this paper, we investigate difficulties and limitations when training networks with reduced texture bias. In particular, we also show that proper evaluation and meaningful comparisons between methods are not trivial. We introduce BiasBed, a testbed for texture- and style-biased training, including multiple datasets and a range of existing algorithms. It comes with an extensive evaluation protocol that includes rigorous hypothesis testing to gauge the significance of the results, despite the considerable training instability of some style bias methods. Our extensive experiments, shed new light on the need for careful, statistically founded evaluation protocols for style bias (and beyond). E.g., we find that some algorithms proposed in the literature do not significantly mitigate the impact of style bias at all. With the release of BiasBed, we hope to foster a common understanding of consistent and meaningful comparisons, and consequently faster progress towards learning methods free of texture bias. Code is available at https://github.com/D1noFuzi/BiasBed
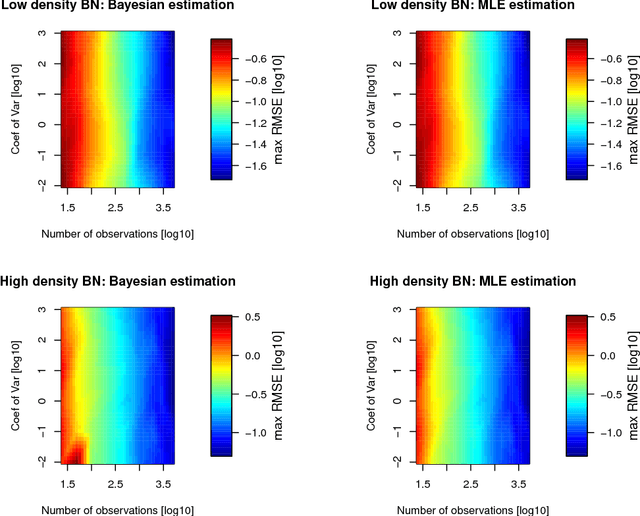
Additive Bayesian Network Modelling with the R Package abn
Nov 20, 2019



Abstract:The R package abn is designed to fit additive Bayesian models to observational datasets. It contains routines to score Bayesian networks based on Bayesian or information theoretic formulations of generalized linear models. It is equipped with exact search and greedy search algorithms to select the best network. It supports a possible blend of continuous, discrete and count data and input of prior knowledge at a structural level. The Bayesian implementation supports random effects to control for one-layer clustering. In this paper, we give an overview of the methodology and illustrate the package's functionalities using a veterinary dataset about respiratory diseases in commercial swine production.
Is a single unique Bayesian network enough to accurately represent your data?
Feb 18, 2019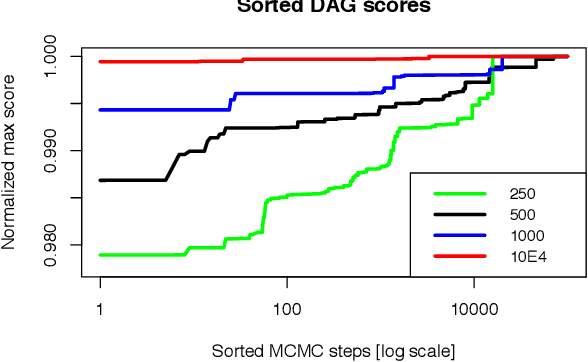
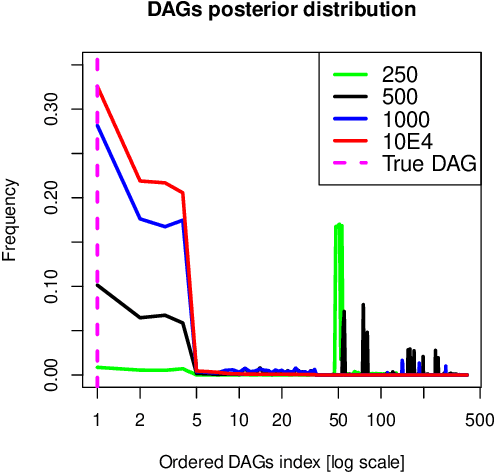
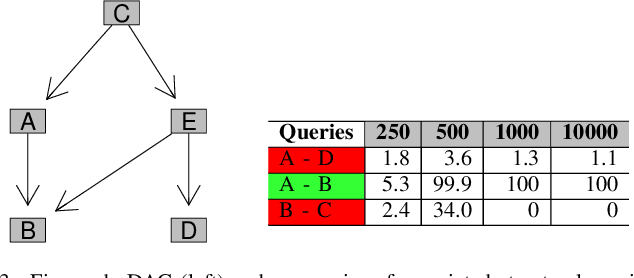
Abstract:Bayesian network (BN) modelling is extensively used in systems epidemiology. Usually it consists in selecting and reporting the best-fitting structure conditional to the data. A major practical concern is avoiding overfitting, on account of its extreme flexibility and its modelling richness. Many approaches have been proposed to control for overfitting. Unfortunately, they essentially all rely on very crude decisions that result in too simplistic approaches for such complex systems. In practice, with limited data sampled from complex system, this approach seems too simplistic. An alternative would be to use the Monte Carlo Markov chain model choice (MC3) over the network to learn the landscape of reasonably supported networks, and then to present all possible arcs with their MCMC support. This paper presents an R implementation, called mcmcabn, of a flexible structural MC3 that is accessible to non-specialists.
Comparison between Suitable Priors for Additive Bayesian Networks
Sep 18, 2018
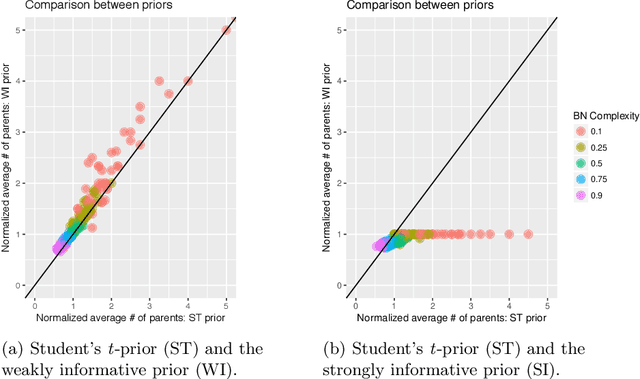
Abstract:Additive Bayesian networks are types of graphical models that extend the usual Bayesian generalized linear model to multiple dependent variables through the factorisation of the joint probability distribution of the underlying variables. When fitting an ABN model, the choice of the prior of the parameters is of crucial importance. If an inadequate prior - like a too weakly informative one - is used, data separation and data sparsity lead to issues in the model selection process. In this work a simulation study between two weakly and a strongly informative priors is presented. As weakly informative prior we use a zero mean Gaussian prior with a large variance, currently implemented in the R-package abn. The second prior belongs to the Student's t-distribution, specifically designed for logistic regressions and, finally, the strongly informative prior is again Gaussian with mean equal to true parameter value and a small variance. We compare the impact of these priors on the accuracy of the learned additive Bayesian network in function of different parameters. We create a simulation study to illustrate Lindley's paradox based on the prior choice. We then conclude by highlighting the good performance of the informative Student's t-prior and the limited impact of the Lindley's paradox. Finally, suggestions for further developments are provided.
Information-Theoretic Scoring Rules to Learn Additive Bayesian Network Applied to Epidemiology
Aug 03, 2018

Abstract:Bayesian network modelling is a well adapted approach to study messy and highly correlated datasets which are very common in, e.g., systems epidemiology. A popular approach to learn a Bayesian network from an observational datasets is to identify the maximum a posteriori network in a search-and-score approach. Many scores have been proposed both Bayesian or frequentist based. In an applied perspective, a suitable approach would allow multiple distributions for the data and is robust enough to run autonomously. A promising framework to compute scores are generalized linear models. Indeed, there exists fast algorithms for estimation and many tailored solutions to common epidemiological issues. The purpose of this paper is to present an R package abn that has an implementation of multiple frequentist scores and some realistic simulations that show its usability and performance. It includes features to deal efficiently with data separation and adjustment which are very common in systems epidemiology.
varrank: an R package for variable ranking based on mutual information with applications to observed systemic datasets
Apr 19, 2018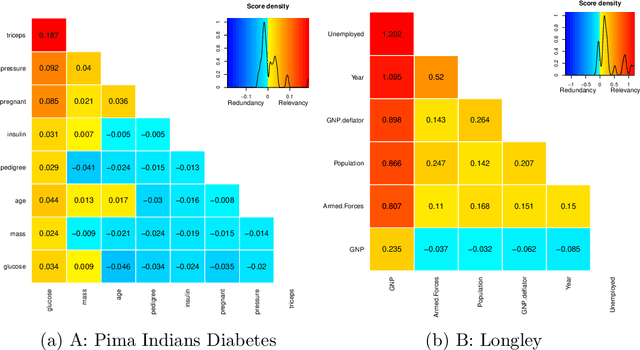
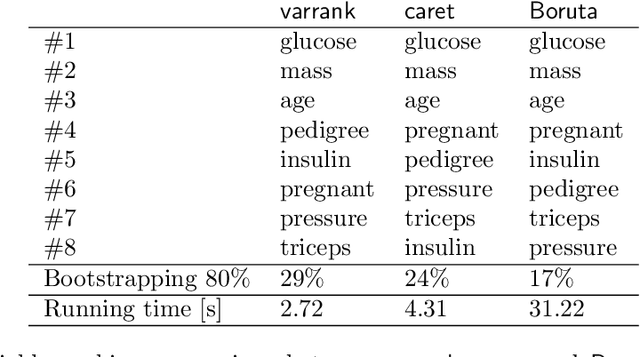
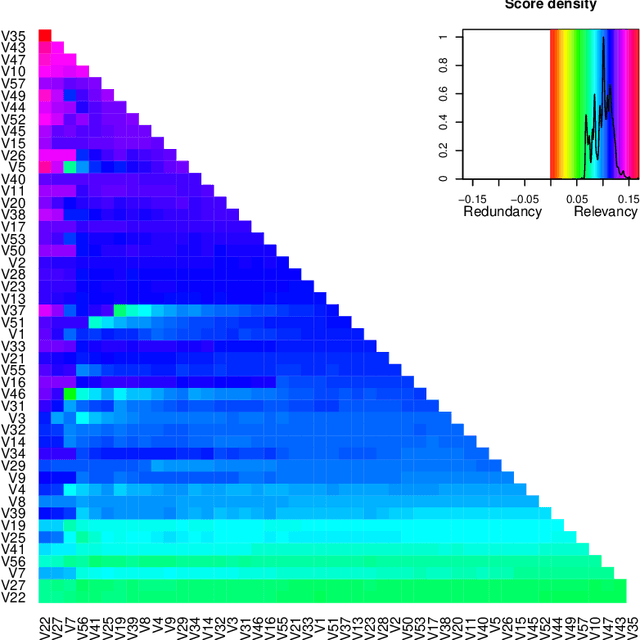
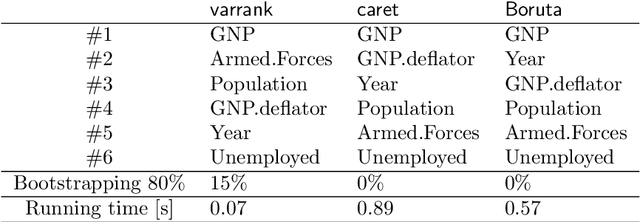
Abstract:This article describes the R package varrank. It has a flexible implementation of heuristic approaches which perform variable ranking based on mutual information. The package is particularly suitable for exploring multivariate datasets requiring a holistic analysis. The core functionality is a general implementation of the minimum redundancy maximum relevance (mRMRe) model. This approach is based on information theory metrics. It is compatible with discrete and continuous data which are discretised using a large choice of possible rules. The two main problems that can be addressed by this package are the selection of the most representative variables for modeling a collection of variables of interest, i.e., dimension reduction, and variable ranking with respect to a set of variables of interest.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge