Preetam Ghosh
GramSeq-DTA: A grammar-based drug-target affinity prediction approach fusing gene expression information
Nov 03, 2024



Abstract:Drug-target affinity (DTA) prediction is a critical aspect of drug discovery. The meaningful representation of drugs and targets is crucial for accurate prediction. Using 1D string-based representations for drugs and targets is a common approach that has demonstrated good results in drug-target affinity prediction. However, these approach lacks information on the relative position of the atoms and bonds. To address this limitation, graph-based representations have been used to some extent. However, solely considering the structural aspect of drugs and targets may be insufficient for accurate DTA prediction. Integrating the functional aspect of these drugs at the genetic level can enhance the prediction capability of the models. To fill this gap, we propose GramSeq-DTA, which integrates chemical perturbation information with the structural information of drugs and targets. We applied a Grammar Variational Autoencoder (GVAE) for drug feature extraction and utilized two different approaches for protein feature extraction: Convolutional Neural Network (CNN) and Recurrent Neural Network (RNN). The chemical perturbation data is obtained from the L1000 project, which provides information on the upregulation and downregulation of genes caused by selected drugs. This chemical perturbation information is processed, and a compact dataset is prepared, serving as the functional feature set of the drugs. By integrating the drug, gene, and target features in the model, our approach outperforms the current state-of-the-art DTA prediction models when validated on widely used DTA datasets (BindingDB, Davis, and KIBA). This work provides a novel and practical approach to DTA prediction by merging the structural and functional aspects of biological entities, and it encourages further research in multi-modal DTA prediction.
A Review of Link Prediction Applications in Network Biology
Dec 03, 2023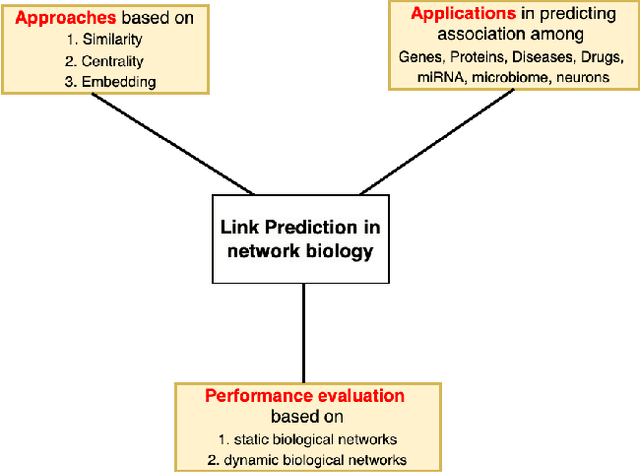
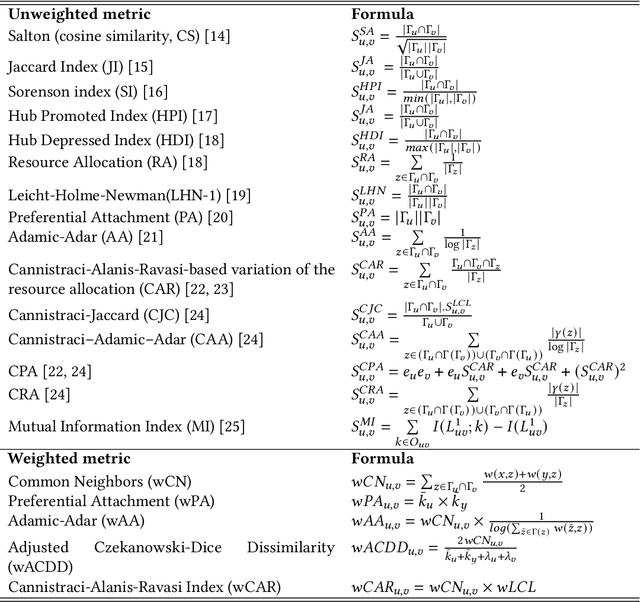
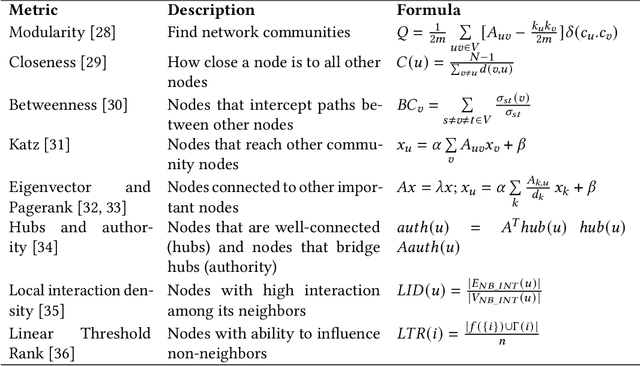
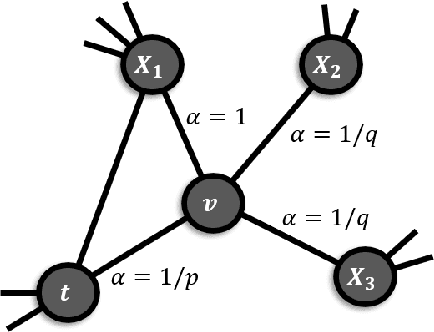
Abstract:In the domain of network biology, the interactions among heterogeneous genomic and molecular entities are represented through networks. Link prediction (LP) methodologies are instrumental in inferring missing or prospective associations within these biological networks. In this review, we systematically dissect the attributes of local, centrality, and embedding-based LP approaches, applied to static and dynamic biological networks. We undertake an examination of the current applications of LP metrics for predicting links between diseases, genes, proteins, RNA, microbiomes, drugs, and neurons. We carry out comprehensive performance evaluations on established biological network datasets to show the practical applications of standard LP models. Moreover, we compare the similarity in prediction trends among the models and the specific network attributes that contribute to effective link prediction, before underscoring the role of LP in addressing the formidable challenges prevalent in biological systems, ranging from noise, bias, and data sparseness to interpretability. We conclude the review with an exploration of the essential characteristics expected from future LP models, poised to advance our comprehension of the intricate interactions governing biological systems.
JobRecoGPT -- Explainable job recommendations using LLMs
Sep 21, 2023



Abstract:In today's rapidly evolving job market, finding the right opportunity can be a daunting challenge. With advancements in the field of AI, computers can now recommend suitable jobs to candidates. However, the task of recommending jobs is not same as recommending movies to viewers. Apart from must-have criteria, like skills and experience, there are many subtle aspects to a job which can decide if it is a good fit or not for a given candidate. Traditional approaches can capture the quantifiable aspects of jobs and candidates, but a substantial portion of the data that is present in unstructured form in the job descriptions and resumes is lost in the process of conversion to structured format. As of late, Large Language Models (LLMs) have taken over the AI field by storm with extraordinary performance in fields where text-based data is available. Inspired by the superior performance of LLMs, we leverage their capability to understand natural language for capturing the information that was previously getting lost during the conversion of unstructured data to structured form. To this end, we compare performance of four different approaches for job recommendations namely, (i) Content based deterministic, (ii) LLM guided, (iii) LLM unguided, and (iv) Hybrid. In this study, we present advantages and limitations of each method and evaluate their performance in terms of time requirements.
Neural Network Architecture for Database Augmentation Using Shared Features
Feb 02, 2023Abstract:The popularity of learning from data with machine learning and neural networks has lead to the creation of many new datasets for almost every problem domain. However, even within a single domain, these datasets are often collected with disparate features, sampled from different sub-populations, and recorded at different time points. Even with the plethora of individual datasets, large data science projects can be difficult as it is often not trivial to merge these smaller datasets. Inherent challenges in some domains such as medicine also makes it very difficult to create large single source datasets or multi-source datasets with identical features. Instead of trying to merge these non-matching datasets directly, we propose a neural network architecture that can provide data augmentation using features common between these datasets. Our results show that this style of data augmentation can work for both image and tabular data.
Deep Hyperspectral Unmixing using Transformer Network
Mar 31, 2022



Abstract:Currently, this paper is under review in IEEE. Transformers have intrigued the vision research community with their state-of-the-art performance in natural language processing. With their superior performance, transformers have found their way in the field of hyperspectral image classification and achieved promising results. In this article, we harness the power of transformers to conquer the task of hyperspectral unmixing and propose a novel deep unmixing model with transformers. We aim to utilize the ability of transformers to better capture the global feature dependencies in order to enhance the quality of the endmember spectra and the abundance maps. The proposed model is a combination of a convolutional autoencoder and a transformer. The hyperspectral data is encoded by the convolutional encoder. The transformer captures long-range dependencies between the representations derived from the encoder. The data are reconstructed using a convolutional decoder. We applied the proposed unmixing model to three widely used unmixing datasets, i.e., Samson, Apex, and Washington DC mall and compared it with the state-of-the-art in terms of root mean squared error and spectral angle distance. The source code for the proposed model will be made publicly available at \url{https://github.com/preetam22n/DeepTrans-HSU}.
Multimodal Classification: Current Landscape, Taxonomy and Future Directions
Sep 18, 2021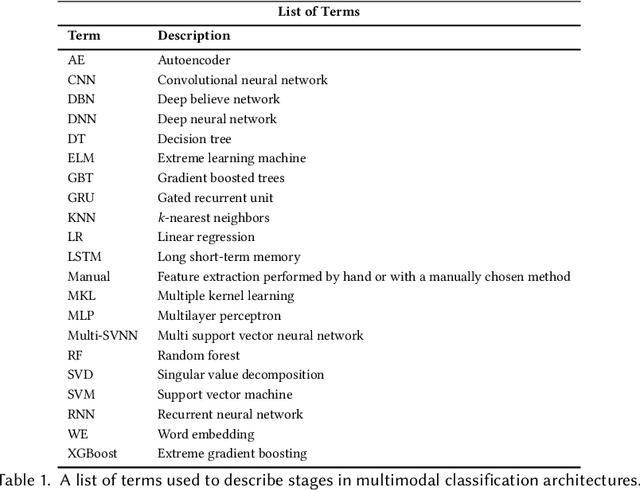
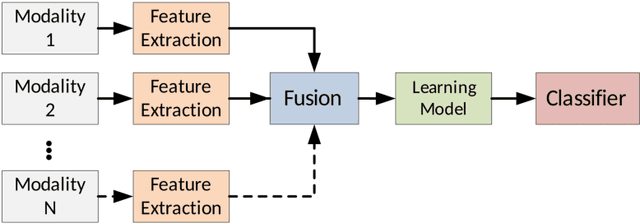
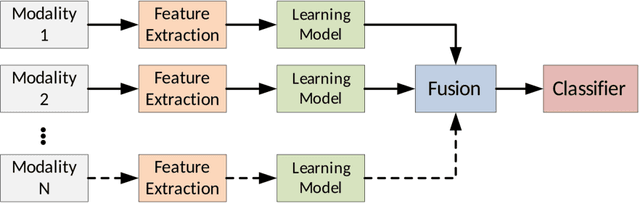
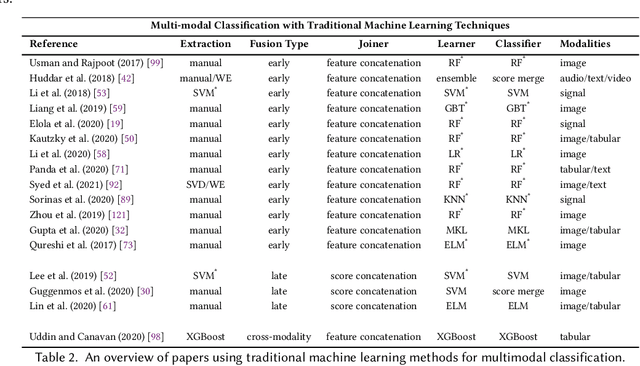
Abstract:Multimodal classification research has been gaining popularity in many domains that collect more data from multiple sources including satellite imagery, biometrics, and medicine. However, the lack of consistent terminology and architectural descriptions makes it difficult to compare different existing solutions. We address these challenges by proposing a new taxonomy for describing such systems based on trends found in recent publications on multimodal classification. Many of the most difficult aspects of unimodal classification have not yet been fully addressed for multimodal datasets including big data, class imbalance, and instance level difficulty. We also provide a discussion of these challenges and future directions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge