Peter B. Gilbert
Inference on Local Variable Importance Measures for Heterogeneous Treatment Effects
Oct 21, 2025Abstract:We provide an inferential framework to assess variable importance for heterogeneous treatment effects. This assessment is especially useful in high-risk domains such as medicine, where decision makers hesitate to rely on black-box treatment recommendation algorithms. The variable importance measures we consider are local in that they may differ across individuals, while the inference is global in that it tests whether a given variable is important for any individual. Our approach builds on recent developments in semiparametric theory for function-valued parameters, and is valid even when statistical machine learning algorithms are employed to quantify treatment effect heterogeneity. We demonstrate the applicability of our method to infectious disease prevention strategies.
On the Role of Surrogates in Conformal Inference of Individual Causal Effects
Dec 16, 2024Abstract:Learning the Individual Treatment Effect (ITE) is essential for personalized decision making, yet causal inference has traditionally focused on aggregated treatment effects. While integrating conformal prediction with causal inference can provide valid uncertainty quantification for ITEs, the resulting prediction intervals are often excessively wide, limiting their practical utility. To address this limitation, we introduce \underline{S}urrogate-assisted \underline{C}onformal \underline{I}nference for \underline{E}fficient I\underline{N}dividual \underline{C}ausal \underline{E}ffects (SCIENCE), a framework designed to construct more efficient prediction intervals for ITEs. SCIENCE applies to various data configurations, including semi-supervised and surrogate-assisted semi-supervised learning. It accommodates covariate shifts between source data, which contain primary outcomes, and target data, which may include only surrogate outcomes or covariates. Leveraging semi-parametric efficiency theory, SCIENCE produces rate double-robust prediction intervals under mild rate convergence conditions, permitting the use of flexible non-parametric models to estimate nuisance functions. We quantify efficiency gains by comparing semi-parametric efficiency bounds with and without the incorporation of surrogates. Simulation studies demonstrate that our surrogate-assisted intervals offer substantial efficiency improvements over existing methods while maintaining valid group-conditional coverage. Applied to the phase 3 Moderna COVE COVID-19 vaccine trial, SCIENCE illustrates how multiple surrogate markers can be leveraged to generate more efficient prediction intervals.
A unified approach for inference on algorithm-agnostic variable importance
Apr 07, 2020
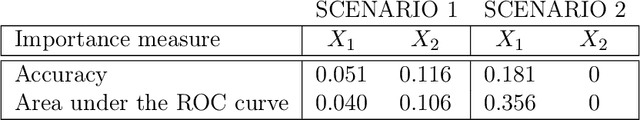
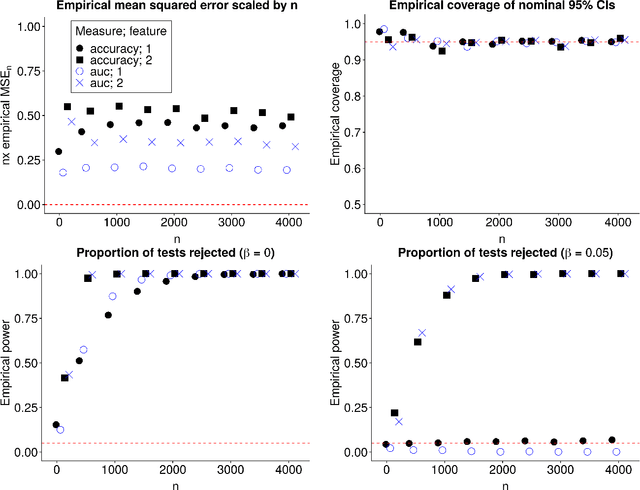

Abstract:In many applications, it is of interest to assess the relative contribution of features (or subsets of features) toward the goal of predicting a response -- in other words, to gauge the variable importance of features. Most recent work on variable importance assessment has focused on describing the importance of features within the confines of a given prediction algorithm. However, such assessment does not necessarily characterize the prediction potential of features, and may provide a misleading reflection of the intrinsic value of these features. To address this limitation, we propose a general framework for nonparametric inference on interpretable algorithm-agnostic variable importance. We define variable importance as a population-level contrast between the oracle predictiveness of all available features versus all features except those under consideration. We propose a nonparametric efficient estimation procedure that allows the construction of valid confidence intervals, even when machine learning techniques are used. We also outline a valid strategy for testing the null importance hypothesis. Through simulations, we show that our proposal has good operating characteristics, and we illustrate its use with data from a study of an antibody against HIV-1 infection.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge