Pericles Philippopoulos
Approximate Network Motif Mining Via Graph Learning
Jun 07, 2022

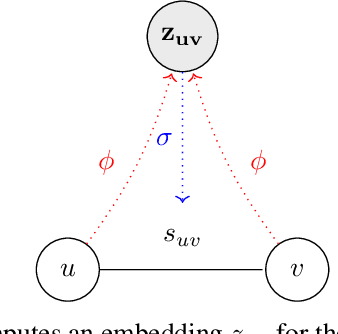
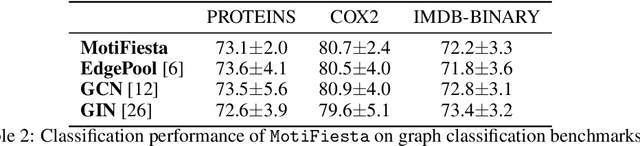
Abstract:Frequent and structurally related subgraphs, also known as network motifs, are valuable features of many graph datasets. However, the high computational complexity of identifying motif sets in arbitrary datasets (motif mining) has limited their use in many real-world datasets. By automatically leveraging statistical properties of datasets, machine learning approaches have shown promise in several tasks with combinatorial complexity and are therefore a promising candidate for network motif mining. In this work we seek to facilitate the development of machine learning approaches aimed at motif mining. We propose a formulation of the motif mining problem as a node labelling task. In addition, we build benchmark datasets and evaluation metrics which test the ability of models to capture different aspects of motif discovery such as motif number, size, topology, and scarcity. Next, we propose MotiFiesta, a first attempt at solving this problem in a fully differentiable manner with promising results on challenging baselines. Finally, we demonstrate through MotiFiesta that this learning setting can be applied simultaneously to general-purpose data mining and interpretable feature extraction for graph classification tasks.
VeRNAl: A Tool for Mining Fuzzy Network Motifs in RNA
Sep 01, 2020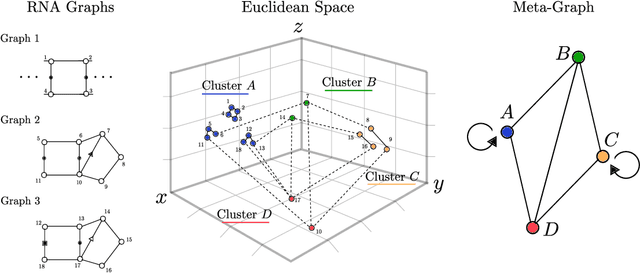


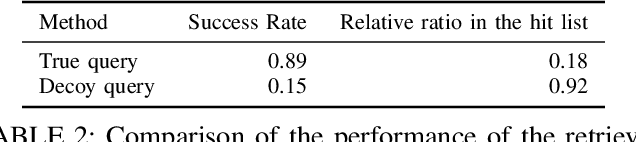
Abstract:Motivation: RNAs are ubiquitous molecules involved in many regulatory and catalytic processes. Their ability to form complex structures is often key to support these functions. Remarkably, RNA 3D structures are articulated around smaller 3D sub-units referred as RNA 3D motifs that can be found in unrelated molecules. The classification of these 3D motifs is thus essential to characterize RNA structures, but current methods can only retrieve motifs with identical base interaction patterns. Results: Here, we relax this constraint by posing the motif finding problem as a graph representation learning and clustering task. This framing takes advantage of the continuous nature of graph representations to model the flexibility of RNA motifs while retaining the convenient encoding of RNAs as graphs. We propose a set of node similarity functions, clustering methods, and motif construction algorithms to recover flexible RNA motifs. We show that our methods are able to retrieve and expand known classes of motifs, but also to identify new motifs. Our tool, VeRNAl can be easily customized by users to desired levels of motif flexibility, abundance and size. Availability and Implementation: The source code, data, and a webserver are available at vernal.cs.mcgill.ca
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge