Orly Reiner
BOrg: A Brain Organoid-Based Mitosis Dataset for Automatic Analysis of Brain Diseases
Jun 27, 2024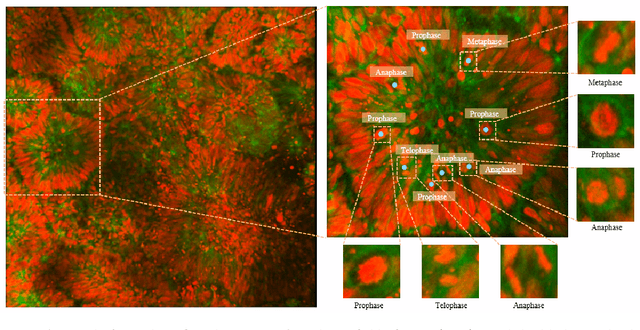
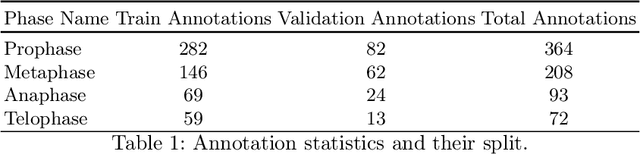
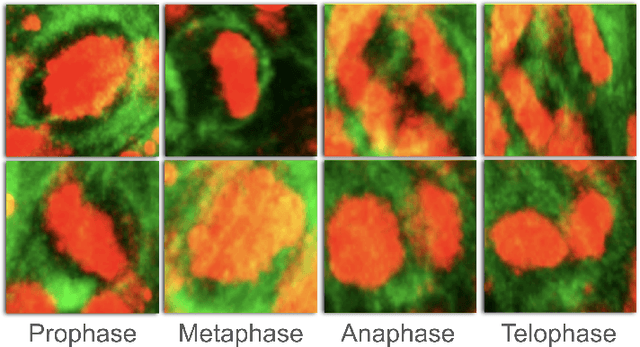
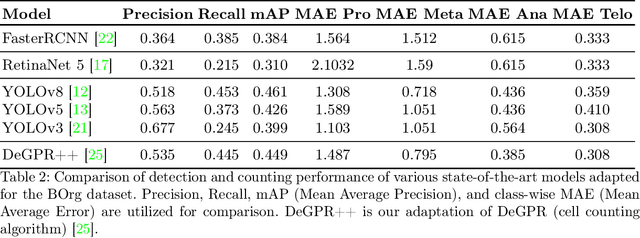
Abstract:Recent advances have enabled the study of human brain development using brain organoids derived from stem cells. Quantifying cellular processes like mitosis in these organoids offers insights into neurodevelopmental disorders, but the manual analysis is time-consuming, and existing datasets lack specific details for brain organoid studies. We introduce BOrg, a dataset designed to study mitotic events in the embryonic development of the brain using confocal microscopy images of brain organoids. BOrg utilizes an efficient annotation pipeline with sparse point annotations and techniques that minimize expert effort, overcoming limitations of standard deep learning approaches on sparse data. We adapt and benchmark state-of-the-art object detection and cell counting models on BOrg for detecting and analyzing mitotic cells across prophase, metaphase, anaphase, and telophase stages. Our results demonstrate these adapted models significantly improve mitosis analysis efficiency and accuracy for brain organoid research compared to existing methods. BOrg facilitates the development of automated tools to quantify statistics like mitosis rates, aiding mechanistic studies of neurodevelopmental processes and disorders. Data and code are available at https://github.com/awaisrauf/borg.
3D Mitochondria Instance Segmentation with Spatio-Temporal Transformers
Mar 21, 2023



Abstract:Accurate 3D mitochondria instance segmentation in electron microscopy (EM) is a challenging problem and serves as a prerequisite to empirically analyze their distributions and morphology. Most existing approaches employ 3D convolutions to obtain representative features. However, these convolution-based approaches struggle to effectively capture long-range dependencies in the volume mitochondria data, due to their limited local receptive field. To address this, we propose a hybrid encoder-decoder framework based on a split spatio-temporal attention module that efficiently computes spatial and temporal self-attentions in parallel, which are later fused through a deformable convolution. Further, we introduce a semantic foreground-background adversarial loss during training that aids in delineating the region of mitochondria instances from the background clutter. Our extensive experiments on three benchmarks, Lucchi, MitoEM-R and MitoEM-H, reveal the benefits of the proposed contributions achieving state-of-the-art results on all three datasets. Our code and models are available at https://github.com/OmkarThawakar/STT-UNET.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge