Onur Afacan
An Optimized Binning and Probabilistic Slice Sharing Algorithm for Motion Correction in Abdominal DW-MRI
Sep 01, 2024Abstract:Abdominal diffusion-weighted magnetic resonance imaging (DW-MRI) is a powerful, non-invasive technique for characterizing lesions and facilitating early diagnosis. However, respiratory motion during a scan can degrade image quality. Binning image slices into respiratory phases may reduce motion artifacts, but when the standard binning algorithm is applied to DW-MRI, reconstructed volumes are often incomplete because they lack slices along the superior-inferior axis. Missing slices create black stripes within images, and prolonged scan times are required to generate complete volumes. In this study, we propose a new binning algorithm to minimize missing slices. We acquired free-breathing and shallow-breathing abdominal DW-MRI scans on seven volunteers and used our algorithm to correct for motion in free-breathing scans. First, we drew the optimal rigid bin partitions in the respiratory signal using a dynamic programming approach, assigning each slice to one bin. We then designed a probabilistic approach for selecting some slices to belong in two bins. Our proposed binning algorithm resulted in significantly fewer missing slices than standard binning (p<1.0e-16), yielding an average reduction of 82.98+/-6.07%. Our algorithm also improved lesion conspicuity and reduced motion artifacts in DW-MR images and Apparent Diffusion Coefficient (ADC) maps. ADC maps created from free-breathing images corrected for motion with our algorithm showed lower intra-subject variability compared to uncorrected free-breathing and shallow-breathing maps (p<0.001). Additionally, shallow-breathing ADC maps showed more consistency with corrected free-breathing maps rather than uncorrected free-breathing maps (p<0.01). Our proposed binning algorithm's efficacy in reducing missing slices increases anatomical accuracy and allows for shorter acquisition times compared to standard binning.
HAITCH: A Framework for Distortion and Motion Correction in Fetal Multi-Shell Diffusion-Weighted MRI
Jun 28, 2024


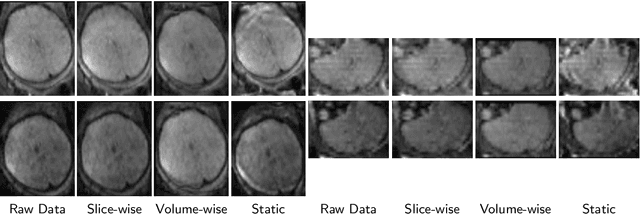
Abstract:Diffusion magnetic resonance imaging (dMRI) is pivotal for probing the microstructure of the rapidly-developing fetal brain. However, fetal motion during scans and its interaction with magnetic field inhomogeneities result in artifacts and data scattering across spatial and angular domains. The effects of those artifacts are more pronounced in high-angular resolution fetal dMRI, where signal-to-noise ratio is very low. Those effects lead to biased estimates and compromise the consistency and reliability of dMRI analysis. This work presents HAITCH, the first and the only publicly available tool to correct and reconstruct multi-shell high-angular resolution fetal dMRI data. HAITCH offers several technical advances that include a blip-reversed dual-echo acquisition for dynamic distortion correction, advanced motion correction for model-free and robust reconstruction, optimized multi-shell design for enhanced information capture and increased tolerance to motion, and outlier detection for improved reconstruction fidelity. The framework is open-source, flexible, and can be used to process any type of fetal dMRI data including single-echo or single-shell acquisitions, but is most effective when used with multi-shell multi-echo fetal dMRI data that cannot be processed with any of the existing tools. Validation experiments on real fetal dMRI scans demonstrate significant improvements and accurate correction across diverse fetal ages and motion levels. HAITCH successfully removes artifacts and reconstructs high-fidelity fetal dMRI data suitable for advanced diffusion modeling, including fiber orientation distribution function estimation. These advancements pave the way for more reliable analysis of the fetal brain microstructure and tractography under challenging imaging conditions.
IVIM-Morph: Motion-compensated quantitative Intra-voxel Incoherent Motion (IVIM) analysis for functional fetal lung maturity assessment from diffusion-weighted MRI data
Jan 17, 2024
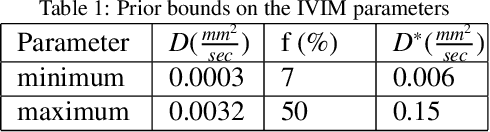

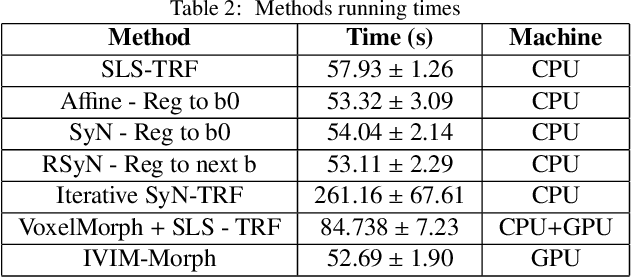
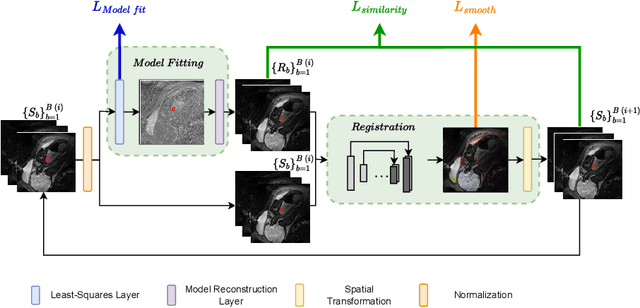
Abstract:Quantitative analysis of pseudo-diffusion in diffusion-weighted magnetic resonance imaging (DWI) data shows potential for assessing fetal lung maturation and generating valuable imaging biomarkers. Yet, the clinical utility of DWI data is hindered by unavoidable fetal motion during acquisition. We present IVIM-morph, a self-supervised deep neural network model for motion-corrected quantitative analysis of DWI data using the Intra-voxel Incoherent Motion (IVIM) model. IVIM-morph combines two sub-networks, a registration sub-network, and an IVIM model fitting sub-network, enabling simultaneous estimation of IVIM model parameters and motion. To promote physically plausible image registration, we introduce a biophysically informed loss function that effectively balances registration and model-fitting quality. We validated the efficacy of IVIM-morph by establishing a correlation between the predicted IVIM model parameters of the lung and gestational age (GA) using fetal DWI data of 39 subjects. IVIM-morph exhibited a notably improved correlation with gestational age (GA) when performing in-vivo quantitative analysis of fetal lung DWI data during the canalicular phase. IVIM-morph shows potential in developing valuable biomarkers for non-invasive assessment of fetal lung maturity with DWI data. Moreover, its adaptability opens the door to potential applications in other clinical contexts where motion compensation is essential for quantitative DWI analysis. The IVIM-morph code is readily available at: https://github.com/TechnionComputationalMRILab/qDWI-Morph.
qDWI-Morph: Motion-compensated quantitative Diffusion-Weighted MRI analysis for fetal lung maturity assessment
Aug 21, 2022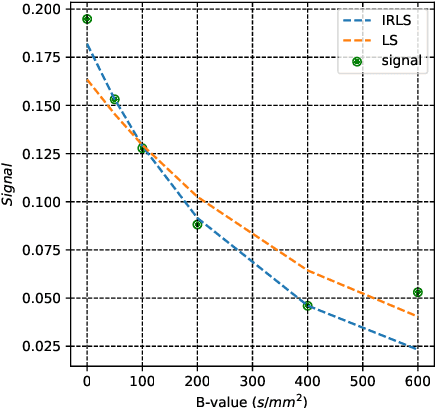

Abstract:Quantitative analysis of fetal lung Diffusion-Weighted MRI (DWI) data shows potential in providing quantitative imaging biomarkers that indirectly reflect fetal lung maturation. However, fetal motion during the acquisition hampered quantitative analysis of the acquired DWI data and, consequently, reliable clinical utilization. We introduce qDWI-morph, an unsupervised deep-neural-network architecture for motion compensated quantitative DWI (qDWI) analysis. Our approach couples a registration sub-network with a quantitative DWI model fitting sub-network. We simultaneously estimate the qDWI parameters and the motion model by minimizing a bio-physically-informed loss function integrating a registration loss and a model fitting quality loss. We demonstrated the added-value of qDWI-morph over: 1) a baseline qDWI analysis without motion compensation and 2) a baseline deep-learning model incorporating registration loss solely. The qDWI-morph achieved a substantially improved correlation with the gestational age through in-vivo qDWI analysis of fetal lung DWI data (R-squared=0.32 vs. 0.13, 0.28). Our qDWI-morph has the potential to enable motion-compensated quantitative analysis of DWI data and to provide clinically feasible bio-markers for non-invasive fetal lung maturity assessment. Our code is available at: https://github.com/TechnionComputationalMRILab/qDWI-Morph.
SUPER-IVIM-DC: Intra-voxel incoherent motion based Fetal lung maturity assessment from limited DWI data using supervised learning coupled with data-consistency
Jun 19, 2022
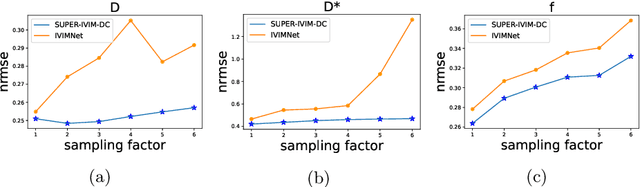
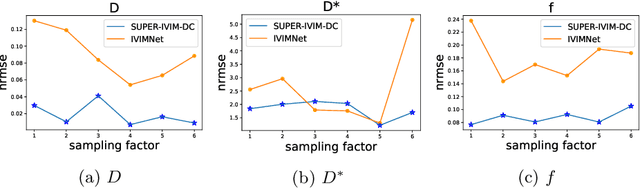
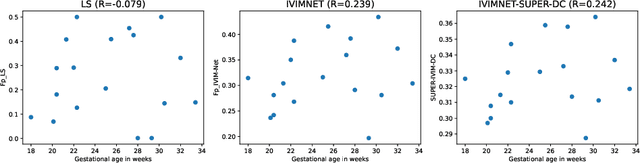
Abstract:Intra-voxel incoherent motion (IVIM) analysis of fetal lungs Diffusion-Weighted MRI (DWI) data shows potential in providing quantitative imaging bio-markers that reflect, indirectly, diffusion and pseudo-diffusion for non-invasive fetal lung maturation assessment. However, long acquisition times, due to the large number of different "b-value" images required for IVIM analysis, precluded clinical feasibility. We introduce SUPER-IVIM-DC a deep-neural-networks (DNN) approach which couples supervised loss with a data-consistency term to enable IVIM analysis of DWI data acquired with a limited number of b-values. We demonstrated the added-value of SUPER-IVIM-DC over both classical and recent DNN approaches for IVIM analysis through numerical simulations, healthy volunteer study, and IVIM analysis of fetal lung maturation from fetal DWI data. Our numerical simulations and healthy volunteer study show that SUPER-IVIM-DC estimates of the IVIM model parameters from limited DWI data had lower normalized root mean-squared error compared to previous DNN-based approaches. Further, SUPER-IVIM-DC estimates of the pseudo-diffusion fraction parameter from limited DWI data of fetal lungs correlate better with gestational age compared to both to classical and DNN-based approaches (0.242 vs. -0.079 and 0.239). SUPER-IVIM-DC has the potential to reduce the long acquisition times associated with IVIM analysis of DWI data and to provide clinically feasible bio-markers for non-invasive fetal lung maturity assessment.
CORPS: Cost-free Rigorous Pseudo-labeling based on Similarity-ranking for Brain MRI Segmentation
May 19, 2022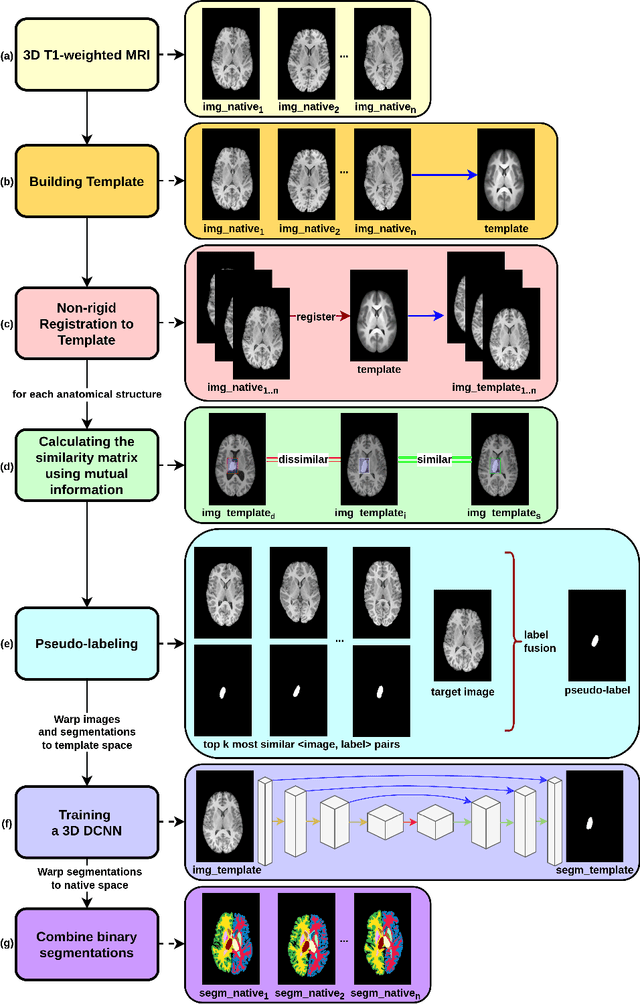
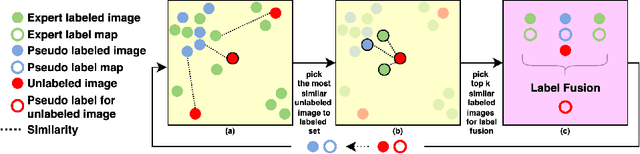
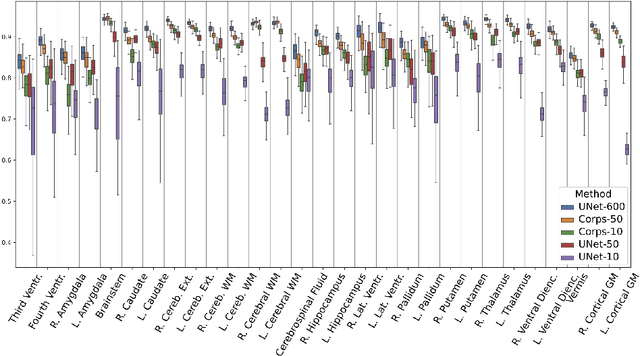
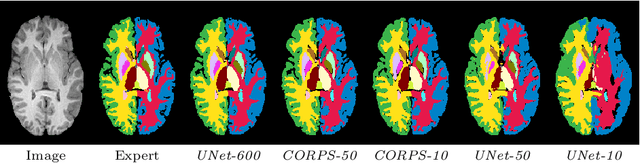
Abstract:Segmentation of brain magnetic resonance images (MRI) is crucial for the analysis of the human brain and diagnosis of various brain disorders. The drawbacks of time-consuming and error-prone manual delineation procedures are aimed to be alleviated by atlas-based and supervised machine learning methods where the former methods are computationally intense and the latter methods lack a sufficiently large number of labeled data. With this motivation, we propose CORPS, a semi-supervised segmentation framework built upon a novel atlas-based pseudo-labeling method and a 3D deep convolutional neural network (DCNN) for 3D brain MRI segmentation. In this work, we propose to generate expert-level pseudo-labels for unlabeled set of images in an order based on a local intensity-based similarity score to existing labeled set of images and using a novel atlas-based label fusion method. Then, we propose to train a 3D DCNN on the combination of expert and pseudo labeled images for binary segmentation of each anatomical structure. The binary segmentation approach is proposed to avoid the poor performance of multi-class segmentation methods on limited and imbalanced data. This also allows to employ a lightweight and efficient 3D DCNN in terms of the number of filters and reserve memory resources for training the binary networks on full-scale and full-resolution 3D MRI volumes instead of 2D/3D patches or 2D slices. Thus, the proposed framework can encapsulate the spatial contiguity in each dimension and enhance context-awareness. The experimental results demonstrate the superiority of the proposed framework over the baseline method both qualitatively and quantitatively without additional labeling cost for manual labeling.
Learning the Regularization in DCE-MR Image Reconstruction for Functional Imaging of Kidneys
Sep 15, 2021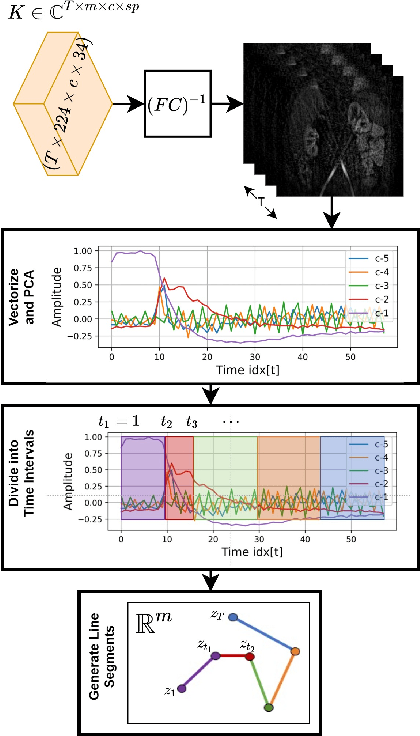
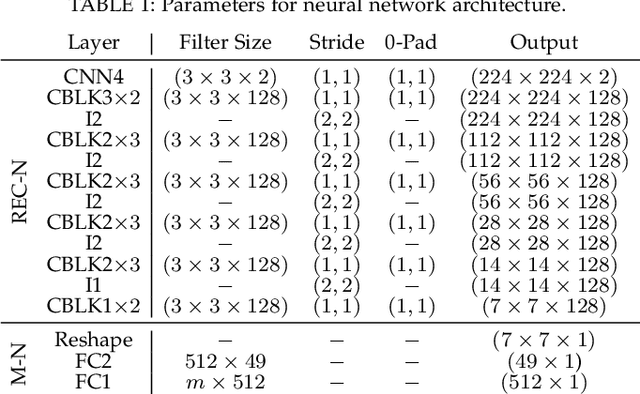
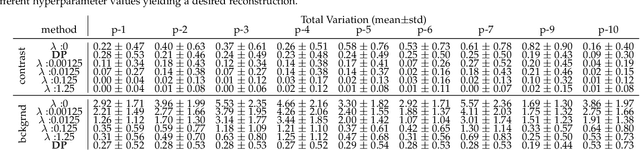
Abstract:Kidney DCE-MRI aims at both qualitative assessment of kidney anatomy and quantitative assessment of kidney function by estimating the tracer kinetic (TK) model parameters. Accurate estimation of TK model parameters requires an accurate measurement of the arterial input function (AIF) with high temporal resolution. Accelerated imaging is used to achieve high temporal resolution, which yields under-sampling artifacts in the reconstructed images. Compressed sensing (CS) methods offer a variety of reconstruction options. Most commonly, sparsity of temporal differences is encouraged for regularization to reduce artifacts. Increasing regularization in CS methods removes the ambient artifacts but also over-smooths the signal temporally which reduces the parameter estimation accuracy. In this work, we propose a single image trained deep neural network to reduce MRI under-sampling artifacts without reducing the accuracy of functional imaging markers. Instead of regularizing with a penalty term in optimization, we promote regularization by generating images from a lower dimensional representation. In this manuscript we motivate and explain the lower dimensional input design. We compare our approach to CS reconstructions with multiple regularization weights. Proposed approach results in kidney biomarkers that are highly correlated with the ground truth markers estimated using the CS reconstruction which was optimized for functional analysis. At the same time, the proposed approach reduces the artifacts in the reconstructed images.
ODE-based Deep Network for MRI Reconstruction
Dec 27, 2019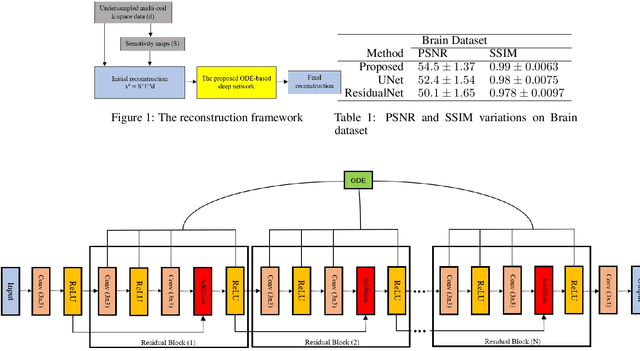
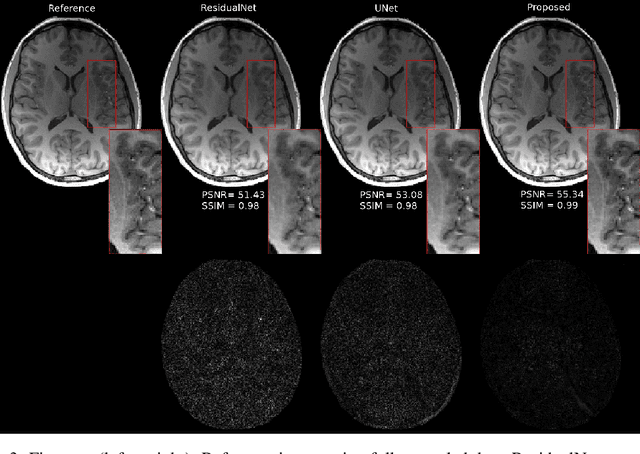
Abstract:Fast data acquisition in Magnetic Resonance Imaging (MRI) is vastly in demand and scan time directly depends on the number of acquired k-space samples. The data-driven methods based on deep neural networks have resulted in promising improvements, compared to the conventional methods, in image reconstruction algorithms. The connection between deep neural network and Ordinary Differential Equation (ODE) has been observed and studied recently. The studies show that different residual networks can be interpreted as Euler discretization of an ODE. In this paper, we propose an ODE-based deep network for MRI reconstruction to enable the rapid acquisition of MR images with improved image quality. Our results with undersampled data demonstrate that our method can deliver higher quality images in comparison to the reconstruction methods based on the standard UNet network and Residual network.
Deep Plug-and-Play Prior for Parallel MRI Reconstruction
Sep 06, 2019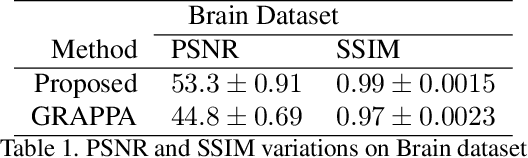
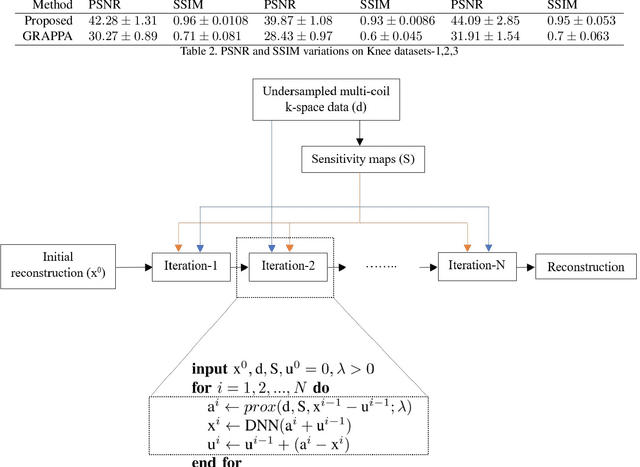
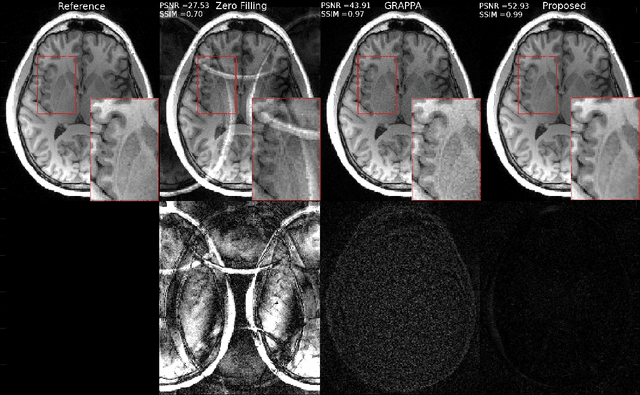
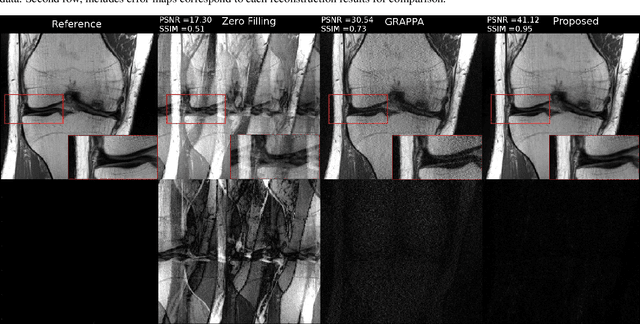
Abstract:Fast data acquisition in Magnetic Resonance Imaging (MRI) is vastly in demand and scan time directly depends on the number of acquired k-space samples. Conventional MRI reconstruction methods for fast MRI acquisition mostly relied on different regularizers which represent analytical models of sparsity. However, recent data-driven methods based on deep learning has resulted in promising improvements in image reconstruction algorithms. In this paper, we propose a deep plug-and-play prior framework for parallel MRI reconstruction problems which utilize a deep neural network (DNN) as an advanced denoiser within an iterative method. This, in turn, enables rapid acquisition of MR images with improved image quality. The proposed method was compared with the reconstructions using the clinical gold standard GRAPPA method. Our results with undersampled data demonstrate that our method can deliver considerably higher quality images at high acceleration factors in comparison to clinical gold standard method for MRI reconstructions. Our proposed reconstruction enables an increase in acceleration factor, and a reduction in acquisition time while maintaining high image quality.
Non-Learning based Deep Parallel MRI Reconstruction (NLDpMRI)
Oct 23, 2018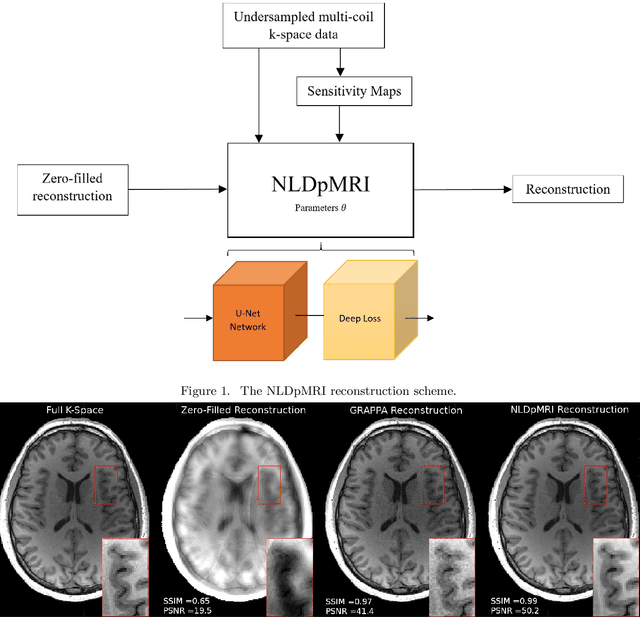
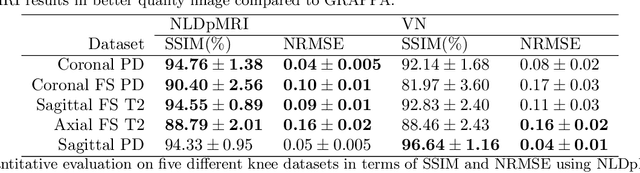
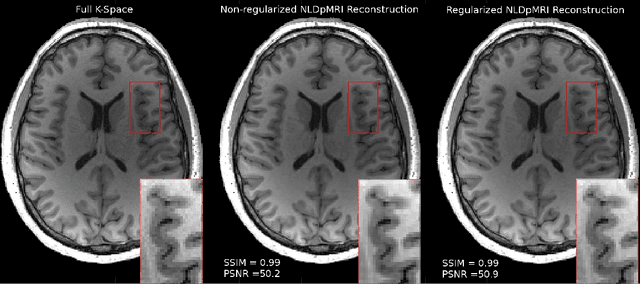
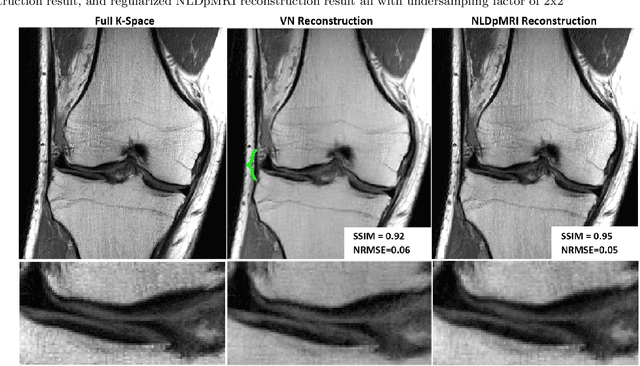
Abstract:Fast data acquisition in Magnetic Resonance Imaging (MRI) is vastly in demand and scan time directly depends on the number of acquired k-space samples. The most common issues in any deep learning-based MRI reconstruction approaches are generalizability and transferability. For different MRI scanner configurations using these approaches, the network must be trained from scratch every time with new training dataset, acquired under new configurations, to be able to provide good reconstruction performance. Here, we propose a new parallel imaging method based on deep neural networks called NLDpMRI to reduce any structured aliasing ambiguities related to the different k-space undersampling patterns for accelerated data acquisition. Two loss functions including non-regularized and regularized are proposed for parallel MRI reconstruction using deep network optimization and we reconstruct MR images by optimizing the proposed loss functions over the network parameters. Unlike any deep learning-based MRI reconstruction approaches, our method doesn't include any training step that the network learns from a large number of training samples and it only needs the single undersampled multi-coil k-space data for reconstruction. Also, the proposed method can handle k-space data with different undersampling patterns, and different number of coils. Unlike most deep learning-based MRI reconstruction methods, our method operates on real-world acquisitions with the complex data format, not on simulated data, real-valued data, or data with added simulated-phase. Experimental results show that the proposed method outperforms the current state-of-the-art GRAPPA reconstruction method.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge