CORPS: Cost-free Rigorous Pseudo-labeling based on Similarity-ranking for Brain MRI Segmentation
Paper and Code
May 19, 2022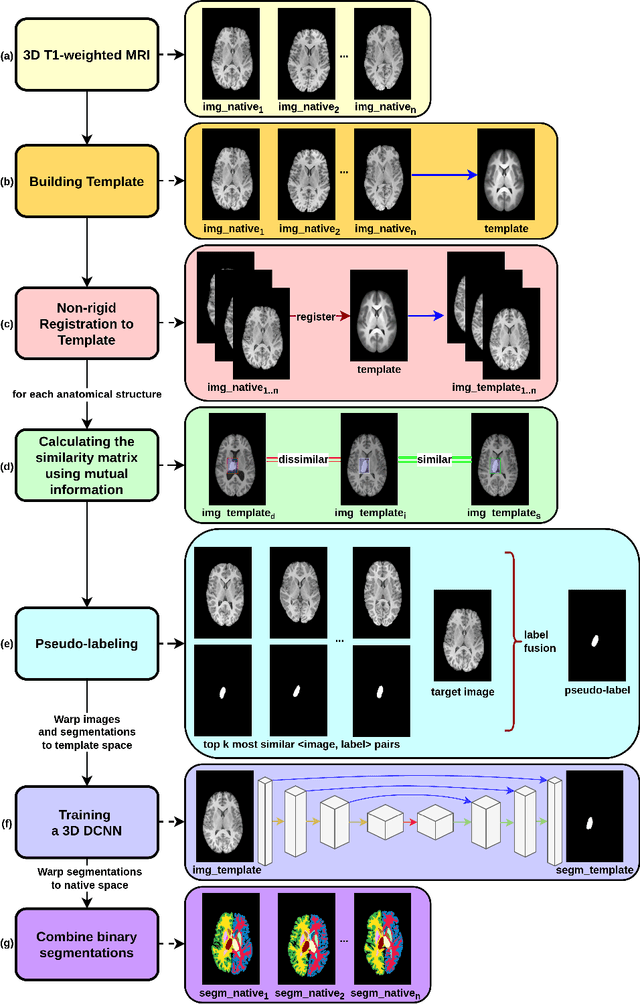
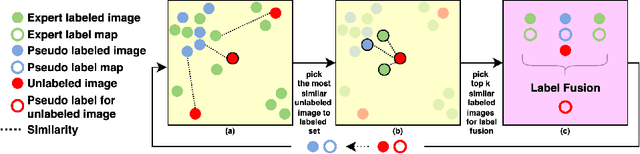
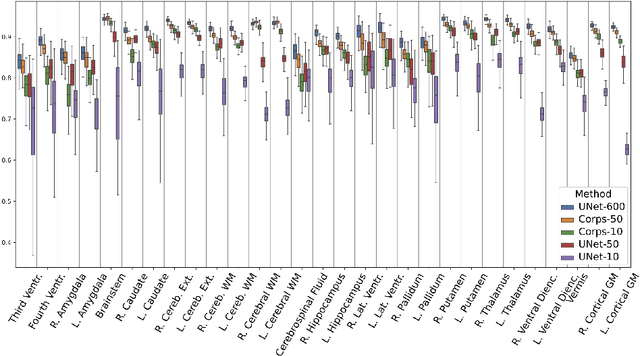
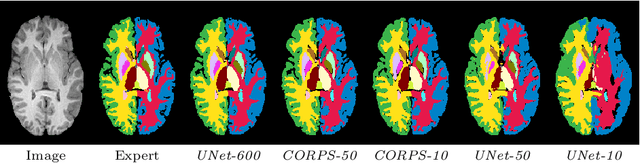
Segmentation of brain magnetic resonance images (MRI) is crucial for the analysis of the human brain and diagnosis of various brain disorders. The drawbacks of time-consuming and error-prone manual delineation procedures are aimed to be alleviated by atlas-based and supervised machine learning methods where the former methods are computationally intense and the latter methods lack a sufficiently large number of labeled data. With this motivation, we propose CORPS, a semi-supervised segmentation framework built upon a novel atlas-based pseudo-labeling method and a 3D deep convolutional neural network (DCNN) for 3D brain MRI segmentation. In this work, we propose to generate expert-level pseudo-labels for unlabeled set of images in an order based on a local intensity-based similarity score to existing labeled set of images and using a novel atlas-based label fusion method. Then, we propose to train a 3D DCNN on the combination of expert and pseudo labeled images for binary segmentation of each anatomical structure. The binary segmentation approach is proposed to avoid the poor performance of multi-class segmentation methods on limited and imbalanced data. This also allows to employ a lightweight and efficient 3D DCNN in terms of the number of filters and reserve memory resources for training the binary networks on full-scale and full-resolution 3D MRI volumes instead of 2D/3D patches or 2D slices. Thus, the proposed framework can encapsulate the spatial contiguity in each dimension and enhance context-awareness. The experimental results demonstrate the superiority of the proposed framework over the baseline method both qualitatively and quantitatively without additional labeling cost for manual labeling.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge