Nhat Khang Ngo
GROOT: Effective Design of Biological Sequences with Limited Experimental Data
Nov 18, 2024



Abstract:Latent space optimization (LSO) is a powerful method for designing discrete, high-dimensional biological sequences that maximize expensive black-box functions, such as wet lab experiments. This is accomplished by learning a latent space from available data and using a surrogate model to guide optimization algorithms toward optimal outputs. However, existing methods struggle when labeled data is limited, as training the surrogate model with few labeled data points can lead to subpar outputs, offering no advantage over the training data itself. We address this challenge by introducing GROOT, a Graph-based Latent Smoothing for Biological Sequence Optimization. In particular, GROOT generates pseudo-labels for neighbors sampled around the training latent embeddings. These pseudo-labels are then refined and smoothed by Label Propagation. Additionally, we theoretically and empirically justify our approach, demonstrate GROOT's ability to extrapolate to regions beyond the training set while maintaining reliability within an upper bound of their expected distances from the training regions. We evaluate GROOT on various biological sequence design tasks, including protein optimization (GFP and AAV) and three tasks with exact oracles from Design-Bench. The results demonstrate that GROOT equalizes and surpasses existing methods without requiring access to black-box oracles or vast amounts of labeled data, highlighting its practicality and effectiveness. We release our code at https://anonymous.4open.science/r/GROOT-D554
Range-aware Positional Encoding via High-order Pretraining: Theory and Practice
Sep 27, 2024Abstract:Unsupervised pre-training on vast amounts of graph data is critical in real-world applications wherein labeled data is limited, such as molecule properties prediction or materials science. Existing approaches pre-train models for specific graph domains, neglecting the inherent connections within networks. This limits their ability to transfer knowledge to various supervised tasks. In this work, we propose a novel pre-training strategy on graphs that focuses on modeling their multi-resolution structural information, allowing us to capture global information of the whole graph while preserving local structures around its nodes. We extend the work of Wave}let Positional Encoding (WavePE) from (Ngo et al., 2023) by pretraining a High-Order Permutation-Equivariant Autoencoder (HOPE-WavePE) to reconstruct node connectivities from their multi-resolution wavelet signals. Unlike existing positional encodings, our method is designed to become sensitivity to the input graph size in downstream tasks, which efficiently capture global structure on graphs. Since our approach relies solely on the graph structure, it is also domain-agnostic and adaptable to datasets from various domains, therefore paving the wave for developing general graph structure encoders and graph foundation models. We theoretically demonstrate that there exists a parametrization of such architecture that it can predict the output adjacency up to arbitrarily low error. We also evaluate HOPE-WavePE on graph-level prediction tasks of different areas and show its superiority compared to other methods.
E(3)-Equivariant Mesh Neural Networks
Feb 19, 2024Abstract:Triangular meshes are widely used to represent three-dimensional objects. As a result, many recent works have address the need for geometric deep learning on 3D mesh. However, we observe that the complexities in many of these architectures does not translate to practical performance, and simple deep models for geometric graphs are competitive in practice. Motivated by this observation, we minimally extend the update equations of E(n)-Equivariant Graph Neural Networks (EGNNs) (Satorras et al., 2021) to incorporate mesh face information, and further improve it to account for long-range interactions through hierarchy. The resulting architecture, Equivariant Mesh Neural Network (EMNN), outperforms other, more complicated equivariant methods on mesh tasks, with a fast run-time and no expensive pre-processing. Our implementation is available at https://github.com/HySonLab/EquiMesh
Multiresolution Graph Transformers and Wavelet Positional Encoding for Learning Hierarchical Structures
Feb 25, 2023



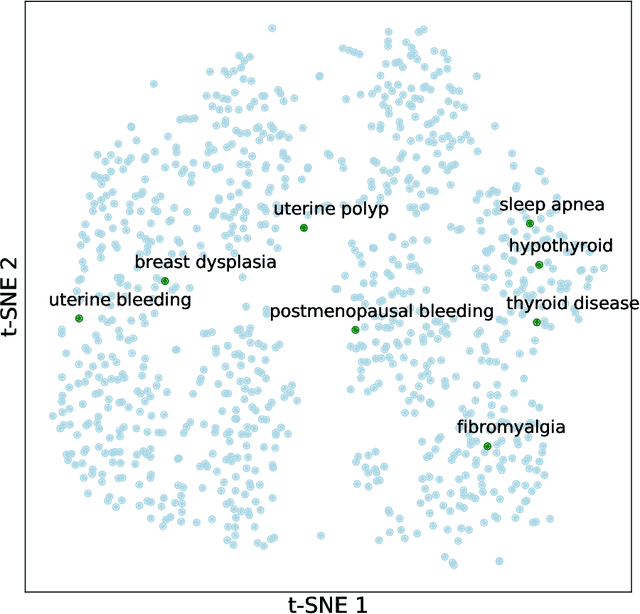
Abstract:Contemporary graph learning algorithms are not well-defined for large molecules since they do not consider the hierarchical interactions among the atoms, which are essential to determine the molecular properties of macromolecules. In this work, we propose Multiresolution Graph Transformers (MGT), the first graph transformer architecture that can learn to represent large molecules at multiple scales. MGT can learn to produce representations for the atoms and group them into meaningful functional groups or repeating units. We also introduce Wavelet Positional Encoding (WavePE), a new positional encoding method that can guarantee localization in both spectral and spatial domains. Our approach achieves competitive results on two macromolecule datasets consisting of polymers and peptides. Furthermore, the visualizations, including clustering results on macromolecules and low-dimensional spaces of their representations, demonstrate the capability of our methodology in learning to represent long-range and hierarchical structures. Our PyTorch implementation is publicly available at https://github.com/HySonLab/Multires-Graph-Transformer.
Modeling Polypharmacy and Predicting Drug-Drug Interactions using Deep Generative Models on Multimodal Graphs
Feb 17, 2023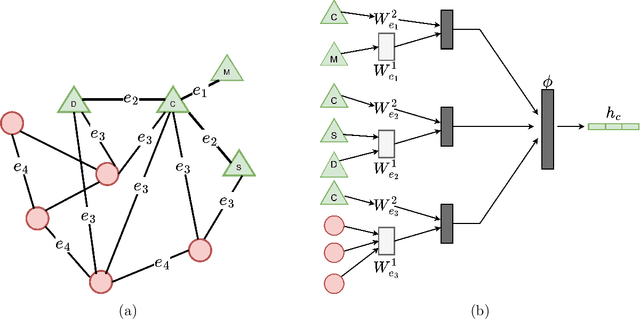
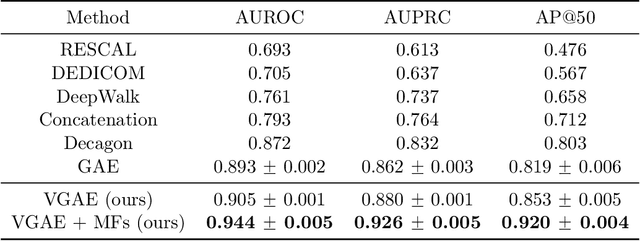
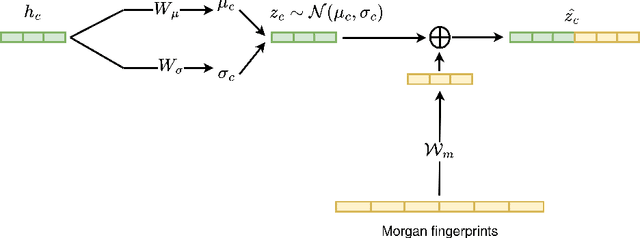
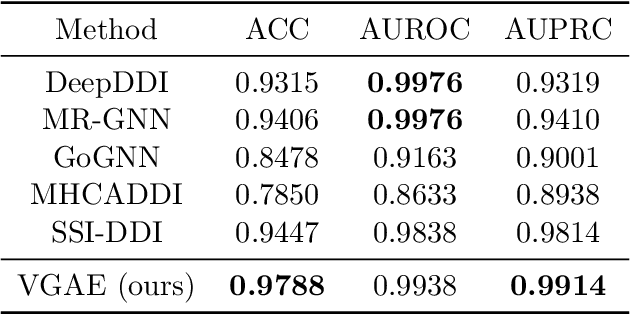
Abstract:Latent representations of drugs and their targets produced by contemporary graph autoencoder models have proved useful in predicting many types of node-pair interactions on large networks, including drug-drug, drug-target, and target-target interactions. However, most existing approaches model either the node's latent spaces in which node distributions are rigid or do not effectively capture the interrelations between drugs; these limitations hinder the methods from accurately predicting drug-pair interactions. In this paper, we present the effectiveness of variational graph autoencoders (VGAE) in modeling latent node representations on multimodal networks. Our approach can produce flexible latent spaces for each node type of the multimodal graph; the embeddings are used later for predicting links among node pairs under different edge types. To further enhance the models' performance, we suggest a new method that concatenates Morgan fingerprints, which capture the molecular structures of each drug, with their latent embeddings before preceding them to the decoding stage for link prediction. Our proposed model shows competitive results on three multimodal networks: (1) a multimodal graph consisting of drug and protein nodes, (2) a multimodal graph constructed from a subset of the DrugBank database involving drug nodes under different interaction types, and (3) a multimodal graph consisting of drug and cell line nodes. Our source code is publicly available at https://github.com/HySonLab/drug-interactions.
Predicting Drug-Drug Interactions using Deep Generative Models on Graphs
Sep 14, 2022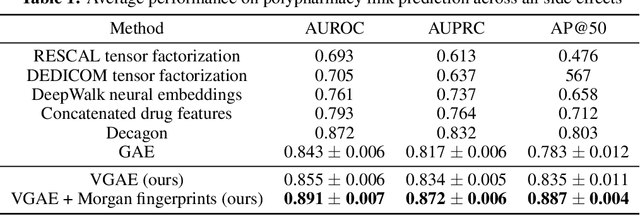


Abstract:Latent representations of drugs and their targets produced by contemporary graph autoencoder-based models have proved useful in predicting many types of node-pair interactions on large networks, including drug-drug, drug-target, and target-target interactions. However, most existing approaches model the node's latent spaces in which node distributions are rigid and disjoint; these limitations hinder the methods from generating new links among pairs of nodes. In this paper, we present the effectiveness of variational graph autoencoders (VGAE) in modeling latent node representations on multimodal networks. Our approach can produce flexible latent spaces for each node type of the multimodal graph; the embeddings are used later for predicting links among node pairs under different edge types. To further enhance the models' performance, we suggest a new method that concatenates Morgan fingerprints, which capture the molecular structures of each drug, with their latent embeddings before preceding them to the decoding stage for link prediction. Our proposed model shows competitive results on two multimodal networks: (1) a multi-graph consisting of drug and protein nodes, and (2) a multi-graph consisting of drug and cell line nodes. Our source code is publicly available at https://github.com/HySonLab/drug-interactions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge