Thanh V. T. Tran
DiFlow-TTS: Discrete Flow Matching with Factorized Speech Tokens for Low-Latency Zero-Shot Text-To-Speech
Sep 11, 2025Abstract:Zero-shot Text-to-Speech (TTS) aims to synthesize high-quality speech that mimics the voice of an unseen speaker using only a short reference sample, requiring not only speaker adaptation but also accurate modeling of prosodic attributes. Recent approaches based on language models, diffusion, and flow matching have shown promising results in zero-shot TTS, but still suffer from slow inference and repetition artifacts. Discrete codec representations have been widely adopted for speech synthesis, and recent works have begun to explore diffusion models in purely discrete settings, suggesting the potential of discrete generative modeling for speech synthesis. However, existing flow-matching methods typically embed these discrete tokens into a continuous space and apply continuous flow matching, which may not fully leverage the advantages of discrete representations. To address these challenges, we introduce DiFlow-TTS, which, to the best of our knowledge, is the first model to explore purely Discrete Flow Matching for speech synthesis. DiFlow-TTS explicitly models factorized speech attributes within a compact and unified architecture. It leverages in-context learning by conditioning on textual content, along with prosodic and acoustic attributes extracted from a reference speech, enabling effective attribute cloning in a zero-shot setting. In addition, the model employs a factorized flow prediction mechanism with distinct heads for prosody and acoustic details, allowing it to learn aspect-specific distributions. Experimental results demonstrate that DiFlow-TTS achieves promising performance in several key metrics, including naturalness, prosody, preservation of speaker style, and energy control. It also maintains a compact model size and achieves low-latency inference, generating speech up to 25.8 times faster than the latest existing baselines.
RESOUND: Speech Reconstruction from Silent Videos via Acoustic-Semantic Decomposed Modeling
May 28, 2025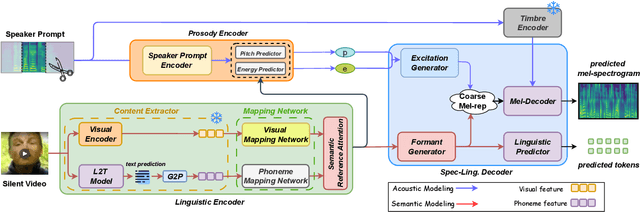
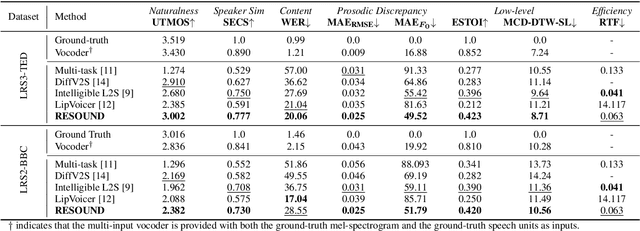

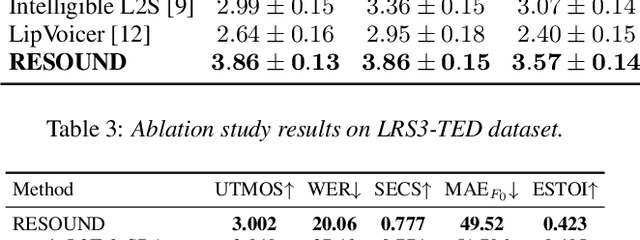
Abstract:Lip-to-speech (L2S) synthesis, which reconstructs speech from visual cues, faces challenges in accuracy and naturalness due to limited supervision in capturing linguistic content, accents, and prosody. In this paper, we propose RESOUND, a novel L2S system that generates intelligible and expressive speech from silent talking face videos. Leveraging source-filter theory, our method involves two components: an acoustic path to predict prosody and a semantic path to extract linguistic features. This separation simplifies learning, allowing independent optimization of each representation. Additionally, we enhance performance by integrating speech units, a proven unsupervised speech representation technique, into waveform generation alongside mel-spectrograms. This allows RESOUND to synthesize prosodic speech while preserving content and speaker identity. Experiments conducted on two standard L2S benchmarks confirm the effectiveness of the proposed method across various metrics.
Effective Context Modeling Framework for Emotion Recognition in Conversations
Dec 21, 2024Abstract:Emotion Recognition in Conversations (ERC) facilitates a deeper understanding of the emotions conveyed by speakers in each utterance within a conversation. Recently, Graph Neural Networks (GNNs) have demonstrated their strengths in capturing data relationships, particularly in contextual information modeling and multimodal fusion. However, existing methods often struggle to fully capture the complex interactions between multiple modalities and conversational context, limiting their expressiveness. To overcome these limitations, we propose ConxGNN, a novel GNN-based framework designed to capture contextual information in conversations. ConxGNN features two key parallel modules: a multi-scale heterogeneous graph that captures the diverse effects of utterances on emotional changes, and a hypergraph that models the multivariate relationships among modalities and utterances. The outputs from these modules are integrated into a fusion layer, where a cross-modal attention mechanism is applied to produce a contextually enriched representation. Additionally, ConxGNN tackles the challenge of recognizing minority or semantically similar emotion classes by incorporating a re-weighting scheme into the loss functions. Experimental results on the IEMOCAP and MELD benchmark datasets demonstrate the effectiveness of our method, achieving state-of-the-art performance compared to previous baselines.
GROOT: Effective Design of Biological Sequences with Limited Experimental Data
Nov 18, 2024



Abstract:Latent space optimization (LSO) is a powerful method for designing discrete, high-dimensional biological sequences that maximize expensive black-box functions, such as wet lab experiments. This is accomplished by learning a latent space from available data and using a surrogate model to guide optimization algorithms toward optimal outputs. However, existing methods struggle when labeled data is limited, as training the surrogate model with few labeled data points can lead to subpar outputs, offering no advantage over the training data itself. We address this challenge by introducing GROOT, a Graph-based Latent Smoothing for Biological Sequence Optimization. In particular, GROOT generates pseudo-labels for neighbors sampled around the training latent embeddings. These pseudo-labels are then refined and smoothed by Label Propagation. Additionally, we theoretically and empirically justify our approach, demonstrate GROOT's ability to extrapolate to regions beyond the training set while maintaining reliability within an upper bound of their expected distances from the training regions. We evaluate GROOT on various biological sequence design tasks, including protein optimization (GFP and AAV) and three tasks with exact oracles from Design-Bench. The results demonstrate that GROOT equalizes and surpasses existing methods without requiring access to black-box oracles or vast amounts of labeled data, highlighting its practicality and effectiveness. We release our code at https://anonymous.4open.science/r/GROOT-D554
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge