Naim U. Rashid
Handling Non-ignorably Missing Features in Electronic Health Records Data Using Importance-Weighted Autoencoders
Feb 05, 2021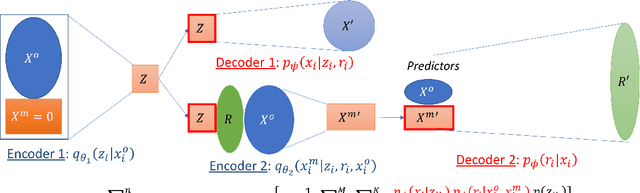
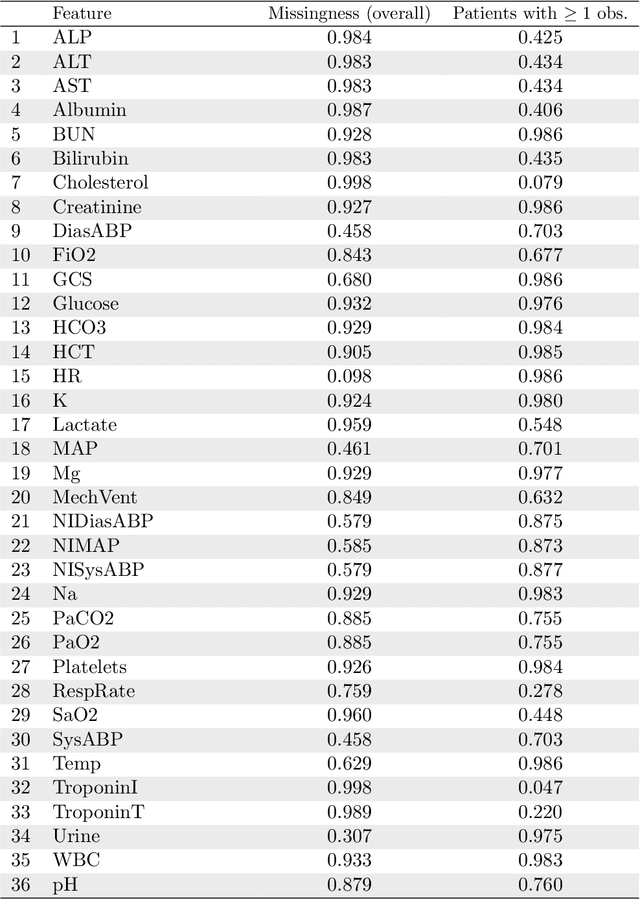
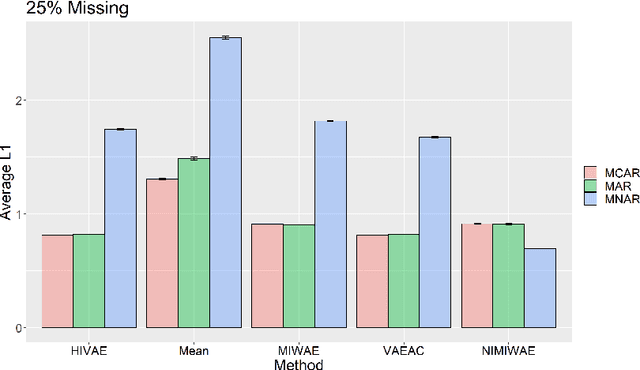
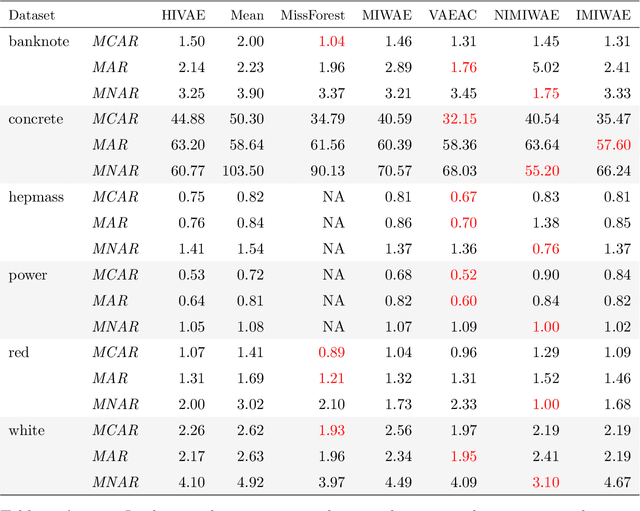
Abstract:Electronic Health Records (EHRs) are commonly used to investigate relationships between patient health information and outcomes. Deep learning methods are emerging as powerful tools to learn such relationships, given the characteristic high dimension and large sample size of EHR datasets. The Physionet 2012 Challenge involves an EHR dataset pertaining to 12,000 ICU patients, where researchers investigated the relationships between clinical measurements, and in-hospital mortality. However, the prevalence and complexity of missing data in the Physionet data present significant challenges for the application of deep learning methods, such as Variational Autoencoders (VAEs). Although a rich literature exists regarding the treatment of missing data in traditional statistical models, it is unclear how this extends to deep learning architectures. To address these issues, we propose a novel extension of VAEs called Importance-Weighted Autoencoders (IWAEs) to flexibly handle Missing Not At Random (MNAR) patterns in the Physionet data. Our proposed method models the missingness mechanism using an embedded neural network, eliminating the need to specify the exact form of the missingness mechanism a priori. We show that the use of our method leads to more realistic imputed values relative to the state-of-the-art, as well as significant differences in fitted downstream models for mortality.
High dimensional precision medicine from patient-derived xenografts
Dec 13, 2019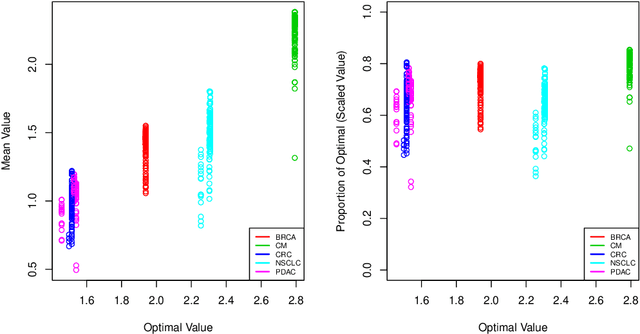

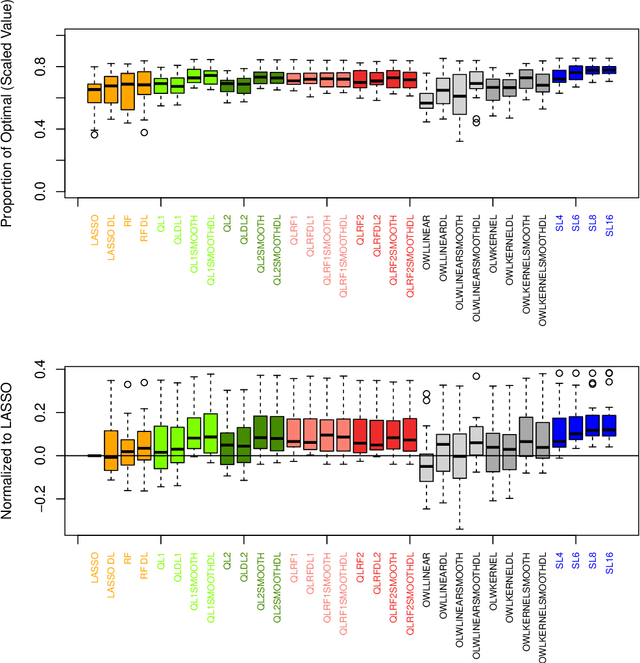
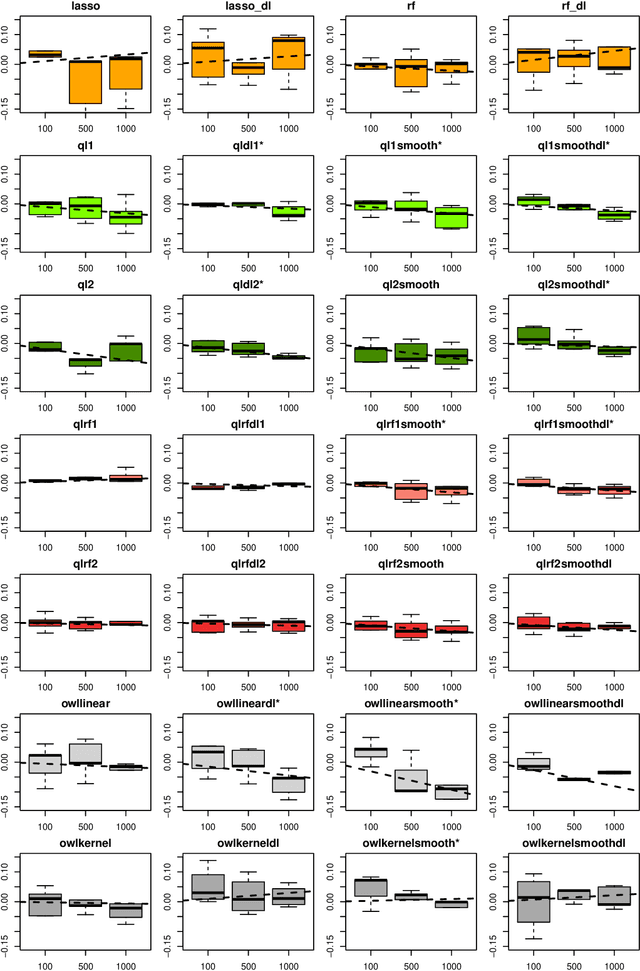
Abstract:The complexity of human cancer often results in significant heterogeneity in response to treatment. Precision medicine offers potential to improve patient outcomes by leveraging this heterogeneity. Individualized treatment rules (ITRs) formalize precision medicine as maps from the patient covariate space into the space of allowable treatments. The optimal ITR is that which maximizes the mean of a clinical outcome in a population of interest. Patient-derived xenograft (PDX) studies permit the evaluation of multiple treatments within a single tumor and thus are ideally suited for estimating optimal ITRs. PDX data are characterized by correlated outcomes, a high-dimensional feature space, and a large number of treatments. Existing methods for estimating optimal ITRs do not take advantage of the unique structure of PDX data or handle the associated challenges well. In this paper, we explore machine learning methods for estimating optimal ITRs from PDX data. We analyze data from a large PDX study to identify biomarkers that are informative for developing personalized treatment recommendations in multiple cancers. We estimate optimal ITRs using regression-based approaches such as Q-learning and direct search methods such as outcome weighted learning. Finally, we implement a superlearner approach to combine a set of estimated ITRs and show that the resulting ITR performs better than any of the input ITRs, mitigating uncertainty regarding user choice of any particular ITR estimation methodology. Our results indicate that PDX data are a valuable resource for developing individualized treatment strategies in oncology.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge