N. Joseph Tatro
Salient Conditional Diffusion for Defending Against Backdoor Attacks
Jan 31, 2023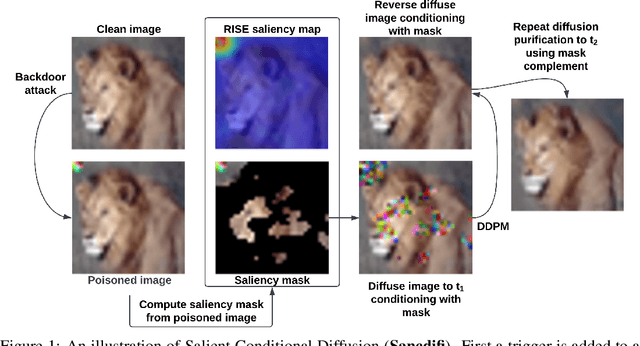
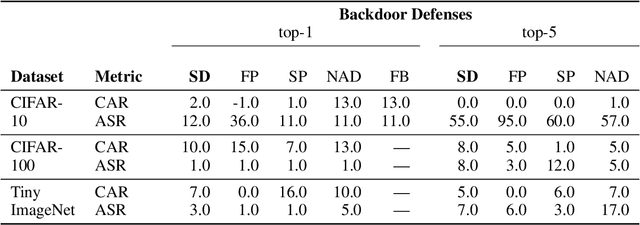
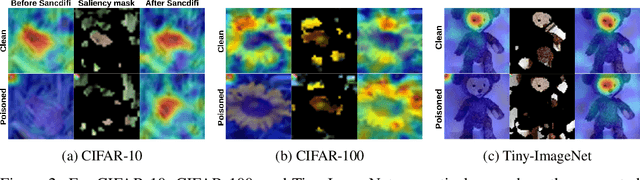
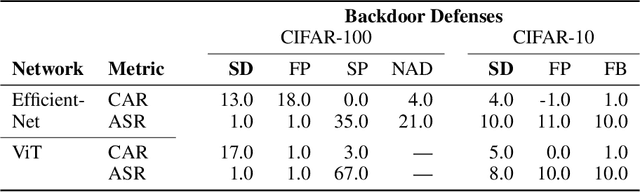
Abstract:We propose a novel algorithm, Salient Conditional Diffusion (Sancdifi), a state-of-the-art defense against backdoor attacks. Sancdifi uses a denoising diffusion probabilistic model (DDPM) to degrade an image with noise and then recover said image using the learned reverse diffusion. Critically, we compute saliency map-based masks to condition our diffusion, allowing for stronger diffusion on the most salient pixels by the DDPM. As a result, Sancdifi is highly effective at diffusing out triggers in data poisoned by backdoor attacks. At the same time, it reliably recovers salient features when applied to clean data. This performance is achieved without requiring access to the model parameters of the Trojan network, meaning Sancdifi operates as a black-box defense.
Learning Geometrically Disentangled Representations of Protein Folding Simulations
May 20, 2022



Abstract:Massive molecular simulations of drug-target proteins have been used as a tool to understand disease mechanism and develop therapeutics. This work focuses on learning a generative neural network on a structural ensemble of a drug-target protein, e.g. SARS-CoV-2 Spike protein, obtained from computationally expensive molecular simulations. Model tasks involve characterizing the distinct structural fluctuations of the protein bound to various drug molecules, as well as efficient generation of protein conformations that can serve as an complement of a molecular simulation engine. Specifically, we present a geometric autoencoder framework to learn separate latent space encodings of the intrinsic and extrinsic geometries of the protein structure. For this purpose, the proposed Protein Geometric AutoEncoder (ProGAE) model is trained on the protein contact map and the orientation of the backbone bonds of the protein. Using ProGAE latent embeddings, we reconstruct and generate the conformational ensemble of a protein at or near the experimental resolution, while gaining better interpretability and controllability in term of protein structure generation from the learned latent space. Additionally, ProGAE models are transferable to a different state of the same protein or to a new protein of different size, where only the dense layer decoding from the latent representation needs to be retrained. Results show that our geometric learning-based method enjoys both accuracy and efficiency for generating complex structural variations, charting the path toward scalable and improved approaches for analyzing and enhancing high-cost simulations of drug-target proteins.
Optimizing Mode Connectivity via Neuron Alignment
Sep 05, 2020



Abstract:The loss landscapes of deep neural networks are not well understood due to their high nonconvexity. Empirically, the local minima of these loss functions can be connected by a learned curve in model space, along which the loss remains nearly constant; a feature known as mode connectivity. Yet, current curve finding algorithms do not consider the influence of symmetry in the loss surface created by model weight permutations. We propose a more general framework to investigate the effect of symmetry on landscape connectivity by accounting for the weight permutations of the networks being connected. To approximate the optimal permutation, we introduce an inexpensive heuristic referred to as neuron alignment. Neuron alignment promotes similarity between the distribution of intermediate activations of models along the curve. We provide theoretical analysis establishing the benefit of alignment to mode connectivity based on this simple heuristic. We empirically verify that the permutation given by alignment is locally optimal via a proximal alternating minimization scheme. Empirically, optimizing the weight permutation is critical for efficiently learning a simple, planar, low-loss curve between networks that successfully generalizes. Our alignment method can significantly alleviate the recently identified robust loss barrier on the path connecting two adversarial robust models and find more robust and accurate models on the path.
Unsupervised Geometric Disentanglement for Surfaces via CFAN-VAE
May 23, 2020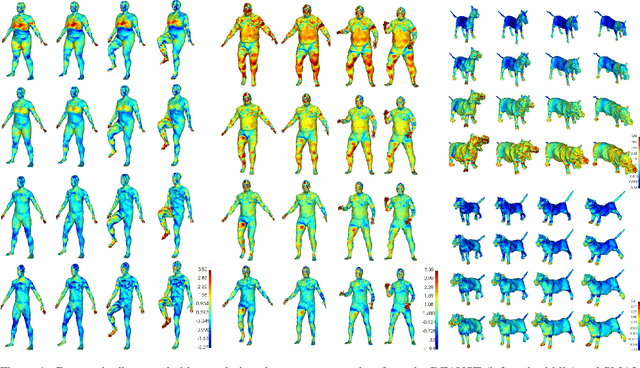
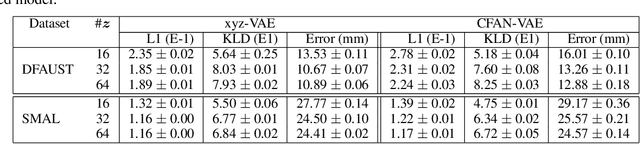
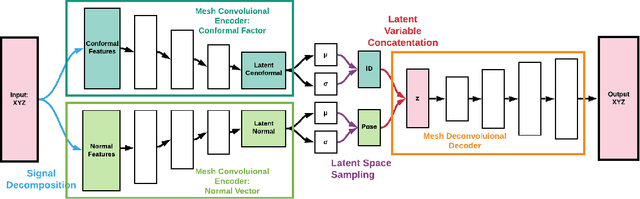
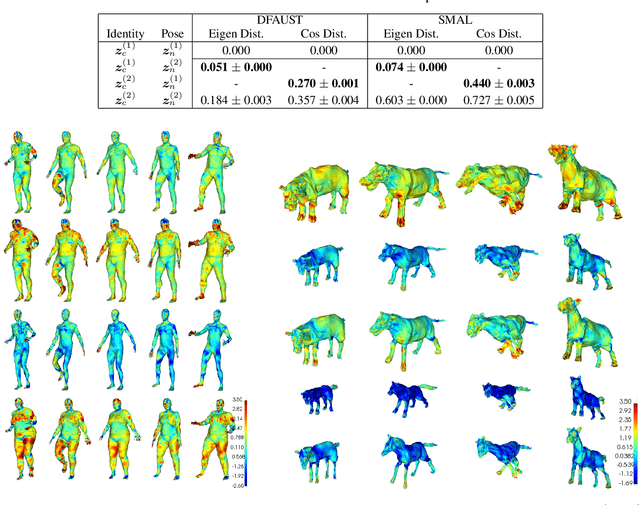
Abstract:For non-Euclidean data such as meshes of humans, a prominent task for generative models is geometric disentanglement; the separation of latent codes for intrinsic (i.e. identity) and extrinsic (i.e. pose) geometry. This work introduces a novel mesh feature, the conformal factor and normal feature (CFAN), for use in mesh convolutional autoencoders. We further propose CFAN-VAE, a novel architecture that disentangles identity and pose using the CFAN feature and parallel transport convolution. CFAN-VAE achieves this geometric disentanglement in an unsupervised way, as it does not require label information on the identity or pose during training. Our comprehensive experiments, including reconstruction, interpolation, generation, and canonical correlation analysis, validate the effectiveness of the unsupervised geometric disentanglement. We also successfully detect and recover geometric disentanglement in mesh convolutional autoencoders that encode xyz-coordinates directly by registering its latent space to that of CFAN-VAE.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge