Morten Grønnesby
A Pragmatic Machine Learning Approach to Quantify Tumor Infiltrating Lymphocytes in Whole Slide Images
Feb 14, 2022
Abstract:Increased levels of tumor infiltrating lymphocytes (TILs) in cancer tissue indicate favourable outcomes in many types of cancer. Manual quantification of immune cells is inaccurate and time consuming for pathologists. Our aim is to leverage a computational solution to automatically quantify TILs in whole slide images (WSIs) of standard diagnostic haematoxylin and eosin stained sections (H&E slides) from lung cancer patients. Our approach is to transfer an open source machine learning method for segmentation and classification of nuclei in H&E slides trained on public data to TIL quantification without manual labeling of our data. Our results show that additional augmentation improves model transferability when training on few samples/limited tissue types. Models trained with sufficient samples/tissue types do not benefit from our additional augmentation policy. Further, the resulting TIL quantification correlates to patient prognosis and compares favorably to the current state-of-the-art method for immune cell detection in non-small lung cancer (current standard CD8 cells in DAB stained TMAs HR 0.34 95% CI 0.17-0.68 vs TILs in HE WSIs: HoVer-Net PanNuke Aug Model HR 0.30 95% CI 0.15-0.60, HoVer-Net MoNuSAC Aug model HR 0.27 95% CI 0.14-0.53). Moreover, we implemented a cloud based system to train, deploy and visually inspect machine learning based annotation for H&E slides. Our pragmatic approach bridges the gap between machine learning research, translational clinical research and clinical implementation. However, validation in prospective studies is needed to assert that the method works in a clinical setting.
Feature Extraction for Machine Learning Based Crackle Detection in Lung Sounds from a Health Survey
Dec 23, 2017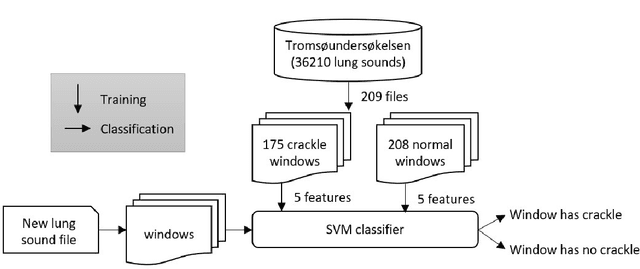
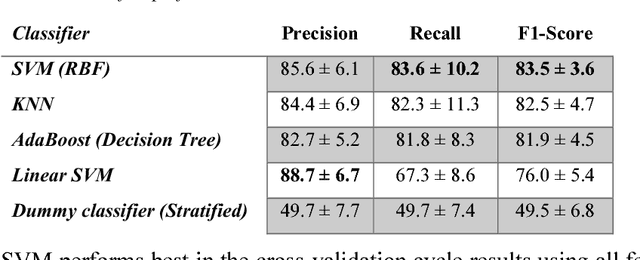
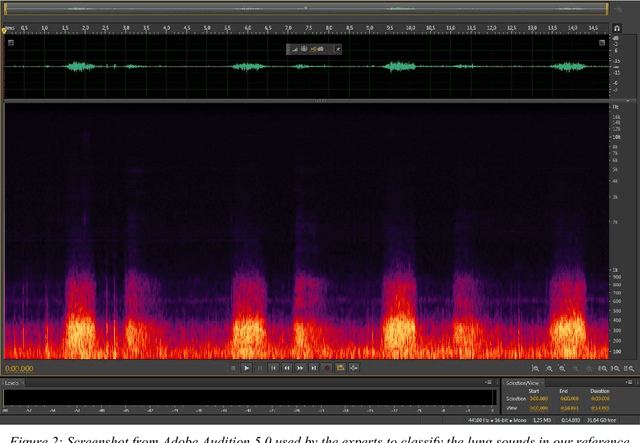
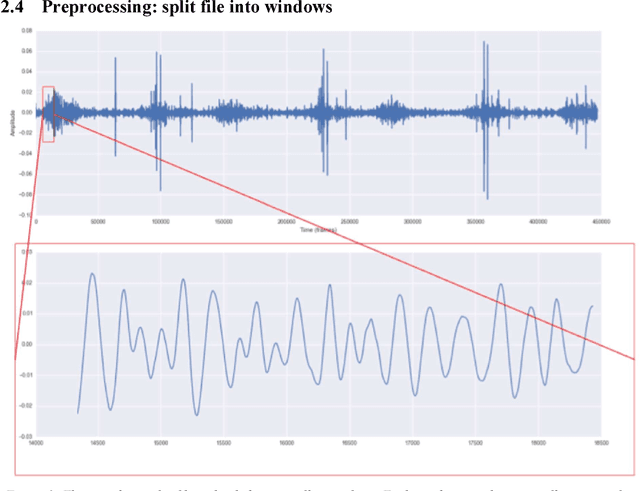
Abstract:In recent years, many innovative solutions for recording and viewing sounds from a stethoscope have become available. However, to fully utilize such devices, there is a need for an automated approach for detecting abnormal lung sounds, which is better than the existing methods that typically have been developed and evaluated using a small and non-diverse dataset. We propose a machine learning based approach for detecting crackles in lung sounds recorded using a stethoscope in a large health survey. Our method is trained and evaluated using 209 files with crackles classified by expert listeners. Our analysis pipeline is based on features extracted from small windows in audio files. We evaluated several feature extraction methods and classifiers. We evaluated the pipeline using a training set of 175 crackle windows and 208 normal windows. We did 100 cycles of cross validation where we shuffled training sets between cycles. For all the division between training and evaluation was 70%-30%. We found and evaluated a 5-dimenstional vector with four features from the time domain and one from the spectrum domain. We evaluated several classifiers and found SVM with a Radial Basis Function Kernel to perform best. Our approach had a precision of 86% and recall of 84% for classifying a crackle in a window, which is more accurate than found in studies of health personnel. The low-dimensional feature vector makes the SVM very fast. The model can be trained on a regular computer in 1.44 seconds, and 319 crackles can be classified in 1.08 seconds. Our approach detects and visualizes individual crackles in recorded audio files. It is accurate, fast, and has low resource requirements. It can be used to train health personnel or as part of a smartphone application for Bluetooth stethoscopes.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge