Edvard Pedersen
Interoperable synthetic health data with SyntHIR to enable the development of CDSS tools
Aug 04, 2023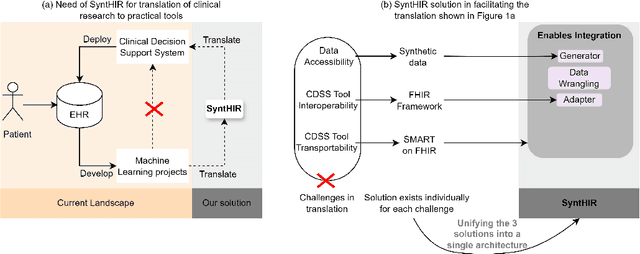

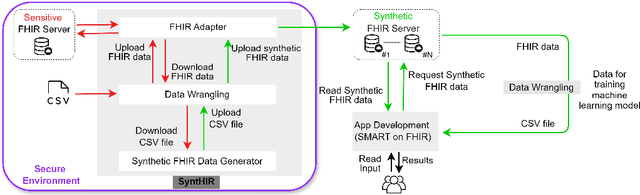
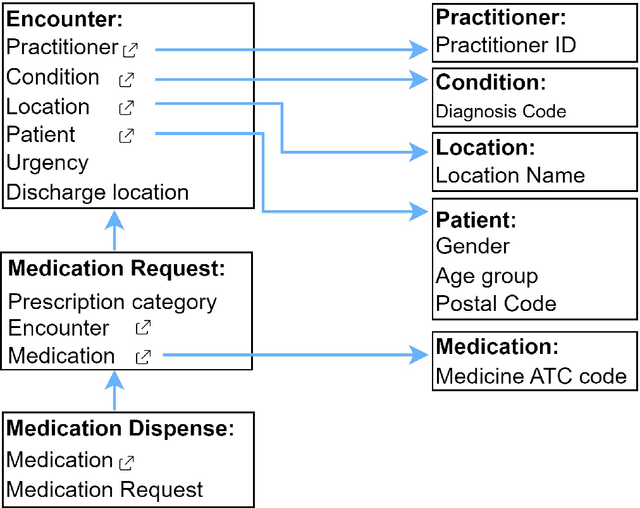
Abstract:There is a great opportunity to use high-quality patient journals and health registers to develop machine learning-based Clinical Decision Support Systems (CDSS). To implement a CDSS tool in a clinical workflow, there is a need to integrate, validate and test this tool on the Electronic Health Record (EHR) systems used to store and manage patient data. However, it is often not possible to get the necessary access to an EHR system due to legal compliance. We propose an architecture for generating and using synthetic EHR data for CDSS tool development. The architecture is implemented in a system called SyntHIR. The SyntHIR system uses the Fast Healthcare Interoperability Resources (FHIR) standards for data interoperability, the Gretel framework for generating synthetic data, the Microsoft Azure FHIR server as the FHIR-based EHR system and SMART on FHIR framework for tool transportability. We demonstrate the usefulness of SyntHIR by developing a machine learning-based CDSS tool using data from the Norwegian Patient Register (NPR) and Norwegian Patient Prescriptions (NorPD). We demonstrate the development of the tool on the SyntHIR system and then lift it to the Open DIPS environment. In conclusion, SyntHIR provides a generic architecture for CDSS tool development using synthetic FHIR data and a testing environment before implementing it in a clinical setting. However, there is scope for improvement in terms of the quality of the synthetic data generated. The code is open source and available at https://github.com/potter-coder89/SyntHIR.git.
A Pragmatic Machine Learning Approach to Quantify Tumor Infiltrating Lymphocytes in Whole Slide Images
Feb 14, 2022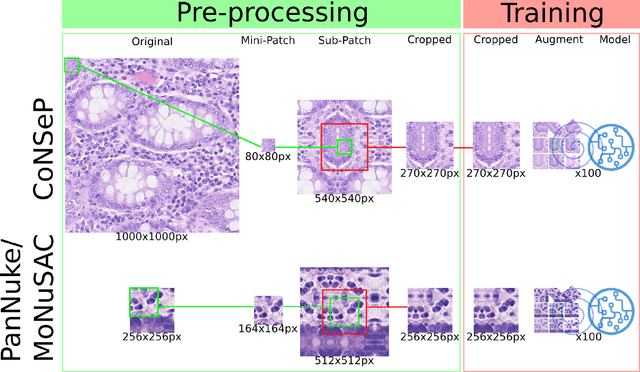
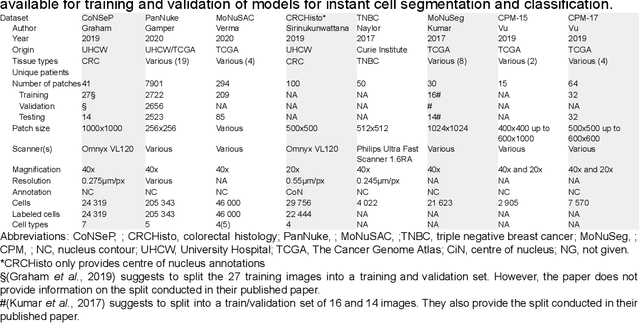
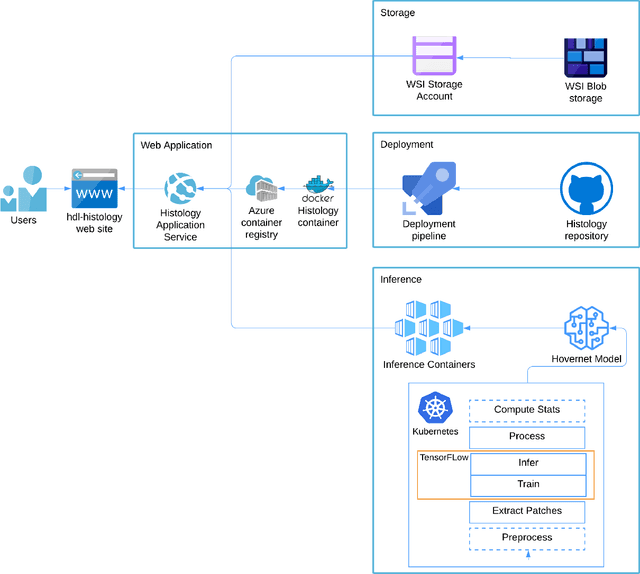
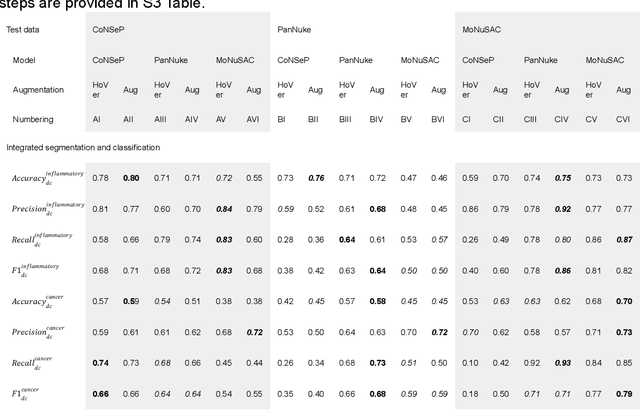
Abstract:Increased levels of tumor infiltrating lymphocytes (TILs) in cancer tissue indicate favourable outcomes in many types of cancer. Manual quantification of immune cells is inaccurate and time consuming for pathologists. Our aim is to leverage a computational solution to automatically quantify TILs in whole slide images (WSIs) of standard diagnostic haematoxylin and eosin stained sections (H&E slides) from lung cancer patients. Our approach is to transfer an open source machine learning method for segmentation and classification of nuclei in H&E slides trained on public data to TIL quantification without manual labeling of our data. Our results show that additional augmentation improves model transferability when training on few samples/limited tissue types. Models trained with sufficient samples/tissue types do not benefit from our additional augmentation policy. Further, the resulting TIL quantification correlates to patient prognosis and compares favorably to the current state-of-the-art method for immune cell detection in non-small lung cancer (current standard CD8 cells in DAB stained TMAs HR 0.34 95% CI 0.17-0.68 vs TILs in HE WSIs: HoVer-Net PanNuke Aug Model HR 0.30 95% CI 0.15-0.60, HoVer-Net MoNuSAC Aug model HR 0.27 95% CI 0.14-0.53). Moreover, we implemented a cloud based system to train, deploy and visually inspect machine learning based annotation for H&E slides. Our pragmatic approach bridges the gap between machine learning research, translational clinical research and clinical implementation. However, validation in prospective studies is needed to assert that the method works in a clinical setting.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge