Thomas K. Kilvaer
A Lightweight and Extensible Cell Segmentation and Classification Model for Whole Slide Images
Feb 26, 2025

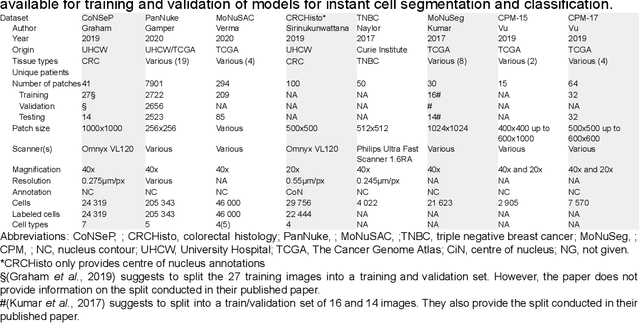
Abstract:Developing clinically useful cell-level analysis tools in digital pathology remains challenging due to limitations in dataset granularity, inconsistent annotations, high computational demands, and difficulties integrating new technologies into workflows. To address these issues, we propose a solution that enhances data quality, model performance, and usability by creating a lightweight, extensible cell segmentation and classification model. First, we update data labels through cross-relabeling to refine annotations of PanNuke and MoNuSAC, producing a unified dataset with seven distinct cell types. Second, we leverage the H-Optimus foundation model as a fixed encoder to improve feature representation for simultaneous segmentation and classification tasks. Third, to address foundation models' computational demands, we distill knowledge to reduce model size and complexity while maintaining comparable performance. Finally, we integrate the distilled model into QuPath, a widely used open-source digital pathology platform. Results demonstrate improved segmentation and classification performance using the H-Optimus-based model compared to a CNN-based model. Specifically, average $R^2$ improved from 0.575 to 0.871, and average $PQ$ score improved from 0.450 to 0.492, indicating better alignment with actual cell counts and enhanced segmentation quality. The distilled model maintains comparable performance while reducing parameter count by a factor of 48. By reducing computational complexity and integrating into workflows, this approach may significantly impact diagnostics, reduce pathologist workload, and improve outcomes. Although the method shows promise, extensive validation is necessary prior to clinical deployment.
Fast TILs estimation in lung cancer WSIs based on semi-stochastic patch sampling
May 05, 2024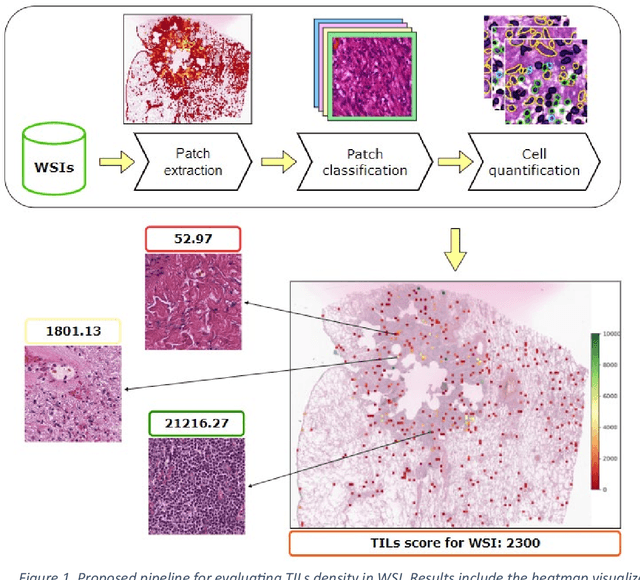
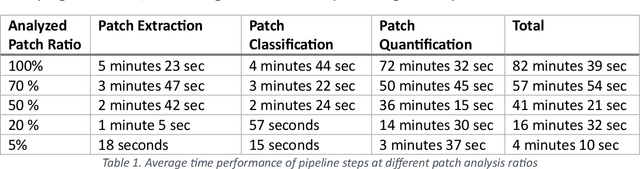
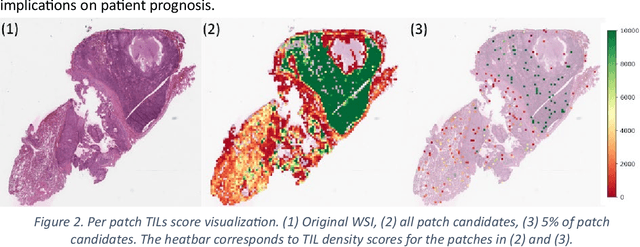
Abstract:Addressing the critical need for accurate prognostic biomarkers in cancer treatment, quantifying tumor-infiltrating lymphocytes (TILs) in non-small cell lung cancer (NSCLC) presents considerable challenges. Manual TIL quantification in whole slide images (WSIs) is laborious and subject to variability, potentially undermining patient outcomes. Our study introduces an automated pipeline that utilizes semi-stochastic patch sampling, patch classification to retain prognostically relevant patches, and cell quantification using the HoVer-Net model to streamline the TIL evaluation process. This pipeline efficiently excludes approximately 70% of areas not relevant for prognosis and requires only 5% of the remaining patches to maintain prognostic accuracy (c-index 0.65 +- 0.01). The computational efficiency achieved does not sacrifice prognostic accuracy, as demonstrated by the TILs score's strong correlation with patient survival, which surpasses traditional CD8 IHC scoring methods. While the pipeline demonstrates potential for enhancing NSCLC prognostication and personalization of treatment, comprehensive clinical validation is still required. Future research should focus on verifying its broader clinical utility and investigating additional biomarkers to improve NSCLC prognosis.
A Pragmatic Machine Learning Approach to Quantify Tumor Infiltrating Lymphocytes in Whole Slide Images
Feb 14, 2022
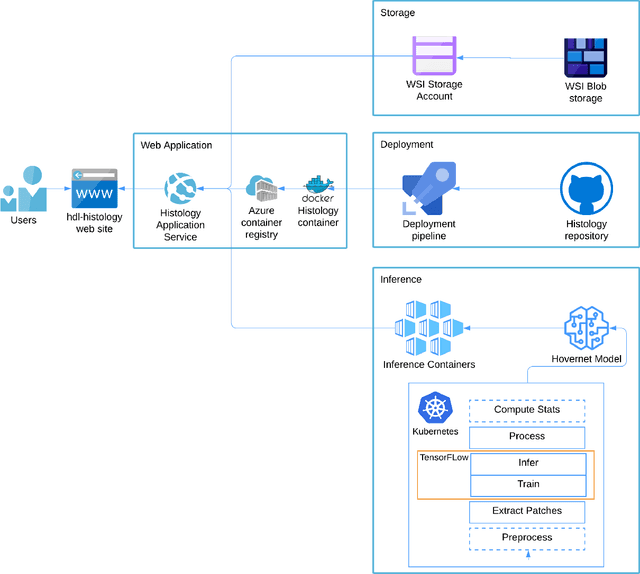
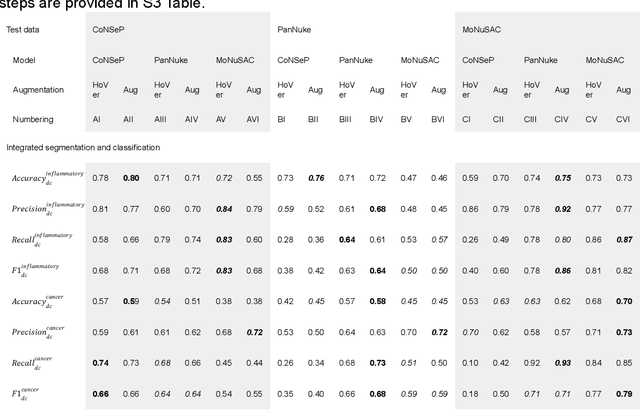
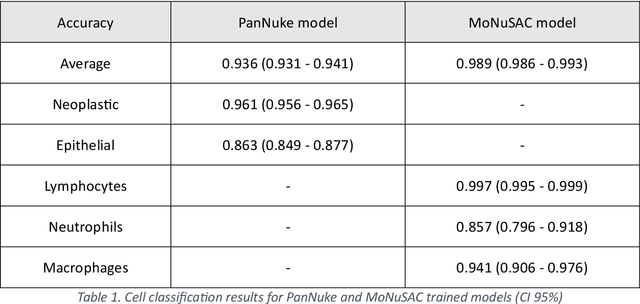
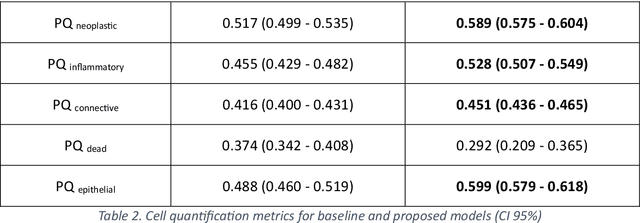
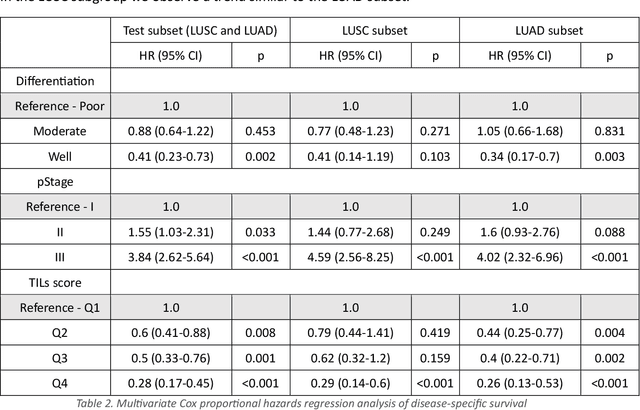
Abstract:Increased levels of tumor infiltrating lymphocytes (TILs) in cancer tissue indicate favourable outcomes in many types of cancer. Manual quantification of immune cells is inaccurate and time consuming for pathologists. Our aim is to leverage a computational solution to automatically quantify TILs in whole slide images (WSIs) of standard diagnostic haematoxylin and eosin stained sections (H&E slides) from lung cancer patients. Our approach is to transfer an open source machine learning method for segmentation and classification of nuclei in H&E slides trained on public data to TIL quantification without manual labeling of our data. Our results show that additional augmentation improves model transferability when training on few samples/limited tissue types. Models trained with sufficient samples/tissue types do not benefit from our additional augmentation policy. Further, the resulting TIL quantification correlates to patient prognosis and compares favorably to the current state-of-the-art method for immune cell detection in non-small lung cancer (current standard CD8 cells in DAB stained TMAs HR 0.34 95% CI 0.17-0.68 vs TILs in HE WSIs: HoVer-Net PanNuke Aug Model HR 0.30 95% CI 0.15-0.60, HoVer-Net MoNuSAC Aug model HR 0.27 95% CI 0.14-0.53). Moreover, we implemented a cloud based system to train, deploy and visually inspect machine learning based annotation for H&E slides. Our pragmatic approach bridges the gap between machine learning research, translational clinical research and clinical implementation. However, validation in prospective studies is needed to assert that the method works in a clinical setting.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge