Minglei Zhao
KDD-LOAM: Jointly Learned Keypoint Detector and Descriptors Assisted LiDAR Odometry and Mapping
Sep 27, 2023



Abstract:Sparse keypoint matching based on distinct 3D feature representations can improve the efficiency and robustness of point cloud registration. Existing learning-based 3D descriptors and keypoint detectors are either independent or loosely coupled, so they cannot fully adapt to each other. In this work, we propose a tightly coupled keypoint detector and descriptor (TCKDD) based on a multi-task fully convolutional network with a probabilistic detection loss. In particular, this self-supervised detection loss fully adapts the keypoint detector to any jointly learned descriptors and benefits the self-supervised learning of descriptors. Extensive experiments on both indoor and outdoor datasets show that our TCKDD achieves state-of-the-art performance in point cloud registration. Furthermore, we design a keypoint detector and descriptors-assisted LiDAR odometry and mapping framework (KDD-LOAM), whose real-time odometry relies on keypoint descriptor matching-based RANSAC. The sparse keypoints are further used for efficient scan-to-map registration and mapping. Experiments on KITTI dataset demonstrate that KDD-LOAM significantly surpasses LOAM and shows competitive performance in odometry.
Artificial Intelligence Advances for De Novo Molecular Structure Modeling in Cryo-EM
Feb 24, 2021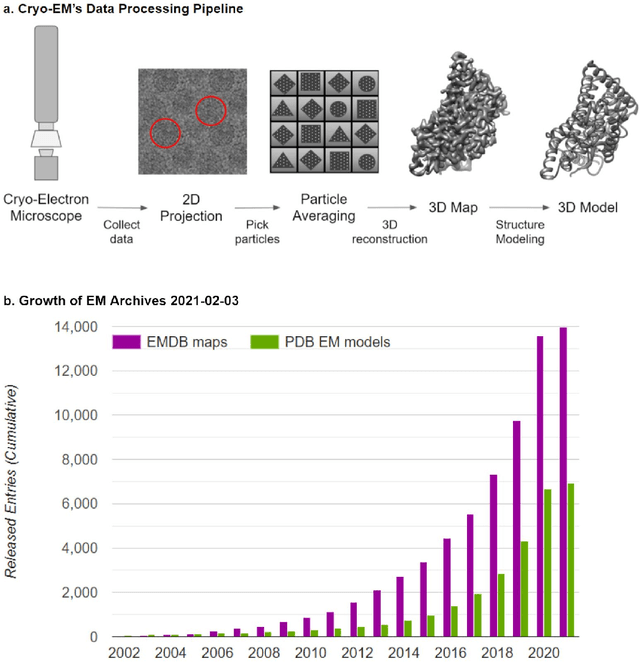
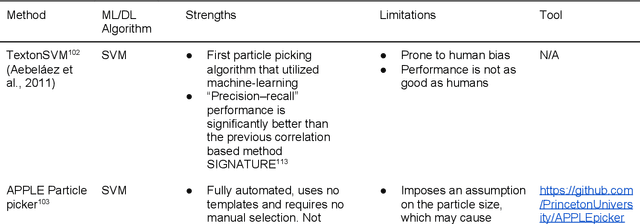
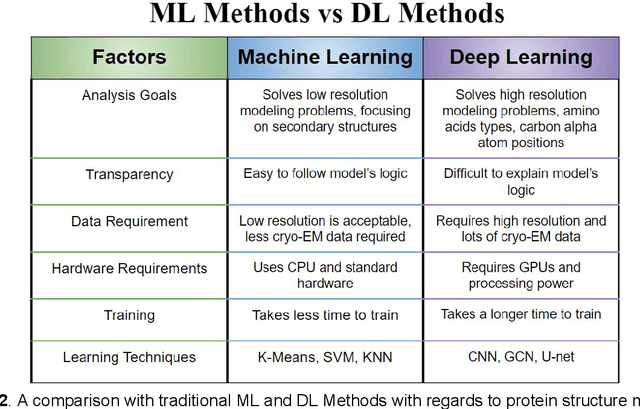
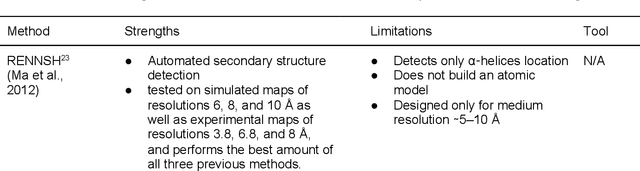
Abstract:Cryo-electron microscopy (cryo-EM) has become a major experimental technique to determine the structures of large protein complexes and molecular assemblies, as evidenced by the 2017 Nobel Prize. Although cryo-EM has been drastically improved to generate high-resolution three-dimensional (3D) maps that contain detailed structural information about macromolecules, the computational methods for using the data to automatically build structure models are lagging far behind. The traditional cryo-EM model building approach is template-based homology modeling. Manual de novo modeling is very time-consuming when no template model is found in the database. In recent years, de novo cryo-EM modeling using machine learning (ML) and deep learning (DL) has ranked among the top-performing methods in macromolecular structure modeling. Deep-learning-based de novo cryo-EM modeling is an important application of artificial intelligence, with impressive results and great potential for the next generation of molecular biomedicine. Accordingly, we systematically review the representative ML/DL-based de novo cryo-EM modeling methods. And their significances are discussed from both practical and methodological viewpoints. We also briefly describe the background of cryo-EM data processing workflow. Overall, this review provides an introductory guide to modern research on artificial intelligence (AI) for de novo molecular structure modeling and future directions in this emerging field.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge