Haowen Guan
Discovering Galaxy Features via Dataset Distillation
Nov 29, 2023Abstract:In many applications, Neural Nets (NNs) have classification performance on par or even exceeding human capacity. Moreover, it is likely that NNs leverage underlying features that might differ from those humans perceive to classify. Can we "reverse-engineer" pertinent features to enhance our scientific understanding? Here, we apply this idea to the notoriously difficult task of galaxy classification: NNs have reached high performance for this task, but what does a neural net (NN) "see" when it classifies galaxies? Are there morphological features that the human eye might overlook that could help with the task and provide new insights? Can we visualize tracers of early evolution, or additionally incorporated spectral data? We present a novel way to summarize and visualize galaxy morphology through the lens of neural networks, leveraging Dataset Distillation, a recent deep-learning methodology with the primary objective to distill knowledge from a large dataset and condense it into a compact synthetic dataset, such that a model trained on this synthetic dataset achieves performance comparable to a model trained on the full dataset. We curate a class-balanced, medium-size high-confidence version of the Galaxy Zoo 2 dataset, and proceed with dataset distillation from our accurate NN-classifier to create synthesized prototypical images of galaxy morphological features, demonstrating its effectiveness. Of independent interest, we introduce a self-adaptive version of the state-of-the-art Matching Trajectory algorithm to automate the distillation process, and show enhanced performance on computer vision benchmarks.
Artificial Intelligence Advances for De Novo Molecular Structure Modeling in Cryo-EM
Feb 24, 2021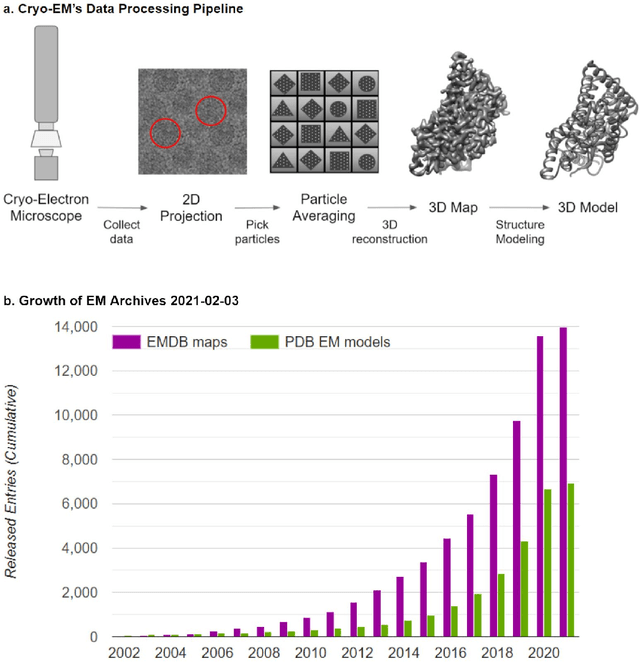
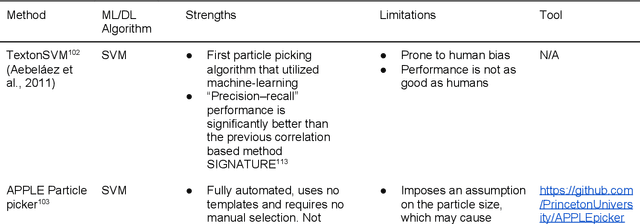
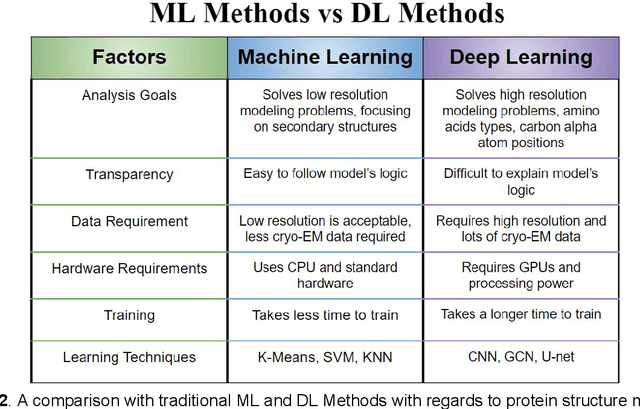
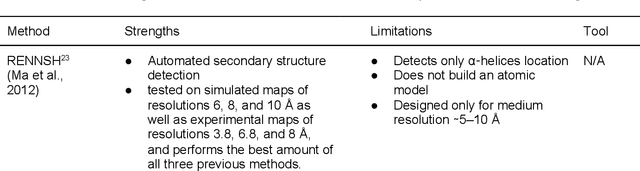
Abstract:Cryo-electron microscopy (cryo-EM) has become a major experimental technique to determine the structures of large protein complexes and molecular assemblies, as evidenced by the 2017 Nobel Prize. Although cryo-EM has been drastically improved to generate high-resolution three-dimensional (3D) maps that contain detailed structural information about macromolecules, the computational methods for using the data to automatically build structure models are lagging far behind. The traditional cryo-EM model building approach is template-based homology modeling. Manual de novo modeling is very time-consuming when no template model is found in the database. In recent years, de novo cryo-EM modeling using machine learning (ML) and deep learning (DL) has ranked among the top-performing methods in macromolecular structure modeling. Deep-learning-based de novo cryo-EM modeling is an important application of artificial intelligence, with impressive results and great potential for the next generation of molecular biomedicine. Accordingly, we systematically review the representative ML/DL-based de novo cryo-EM modeling methods. And their significances are discussed from both practical and methodological viewpoints. We also briefly describe the background of cryo-EM data processing workflow. Overall, this review provides an introductory guide to modern research on artificial intelligence (AI) for de novo molecular structure modeling and future directions in this emerging field.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge