Michael O'Neill
Amplifying Limitations, Harms and Risks of Large Language Models
Jul 06, 2023Abstract:We present this article as a small gesture in an attempt to counter what appears to be exponentially growing hype around Artificial Intelligence (AI) and its capabilities, and the distraction provided by the associated talk of science-fiction scenarios that might arise if AI should become sentient and super-intelligent. It may also help those outside of the field to become more informed about some of the limitations of AI technology. In the current context of popular discourse AI defaults to mean foundation and large language models (LLMs) such as those used to create ChatGPT. This in itself is a misrepresentation of the diversity, depth and volume of research, researchers, and technology that truly represents the field of AI. AI being a field of research that has existed in software artefacts since at least the 1950's. We set out to highlight a number of limitations of LLMs, and in so doing highlight that harms have already arisen and will continue to arise due to these limitations. Along the way we also highlight some of the associated risks for individuals and organisations in using this technology.
Large Language Models in Sport Science & Medicine: Opportunities, Risks and Considerations
May 05, 2023
Abstract:This paper explores the potential opportunities, risks, and challenges associated with the use of large language models (LLMs) in sports science and medicine. LLMs are large neural networks with transformer style architectures trained on vast amounts of textual data, and typically refined with human feedback. LLMs can perform a large range of natural language processing tasks. In sports science and medicine, LLMs have the potential to support and augment the knowledge of sports medicine practitioners, make recommendations for personalised training programs, and potentially distribute high-quality information to practitioners in developing countries. However, there are also potential risks associated with the use and development of LLMs, including biases in the dataset used to create the model, the risk of exposing confidential data, the risk of generating harmful output, and the need to align these models with human preferences through feedback. Further research is needed to fully understand the potential applications of LLMs in sports science and medicine and to ensure that their use is ethical and beneficial to athletes, clients, patients, practitioners, and the general public.
An exploration of asocial and social learning in the evolution of variable-length structures
Apr 16, 2021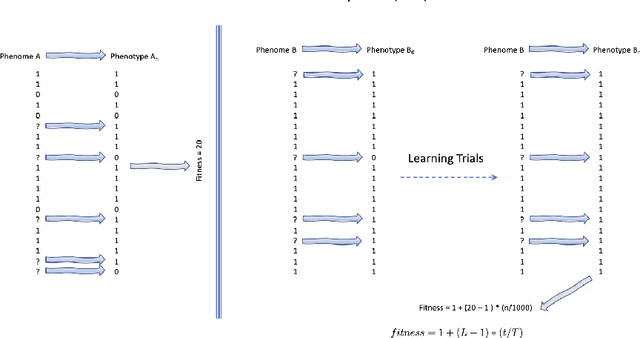
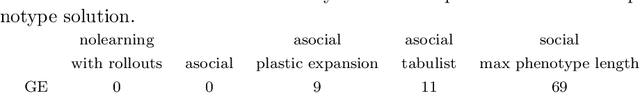
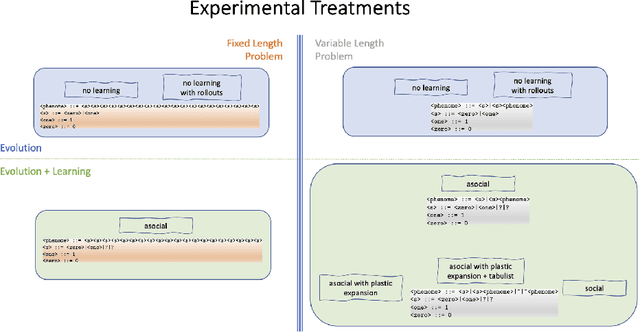
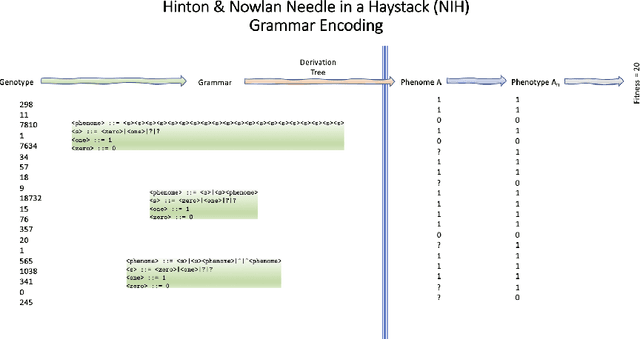
Abstract:We wish to explore the contribution that asocial and social learning might play as a mechanism for self-adaptation in the search for variable-length structures by an evolutionary algorithm. An extremely challenging, yet simple to understand problem landscape is adopted where the probability of randomly finding a solution is approximately one in a trillion. A number of learning mechanisms operating on variable-length structures are implemented and their performance analysed. The social learning setup, which combines forms of both social and asocial learning in combination with evolution is found to be most performant, while the setups exclusively adopting evolution are incapable of finding solutions.
Optimizing the Parameters of A Physical Exercise Dose-Response Model: An Algorithmic Comparison
Dec 16, 2020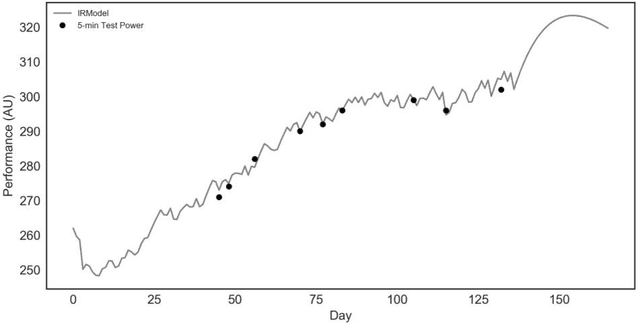
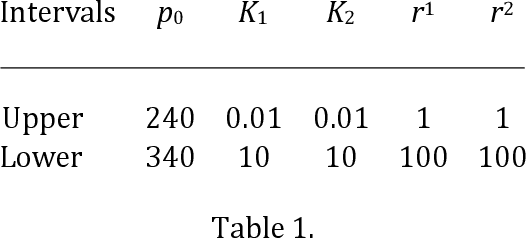
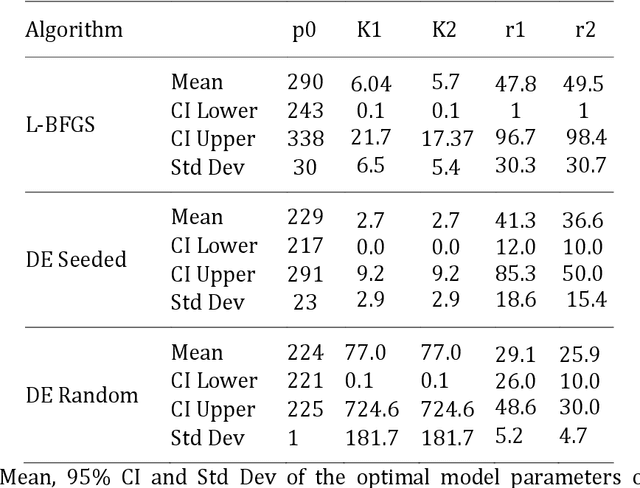
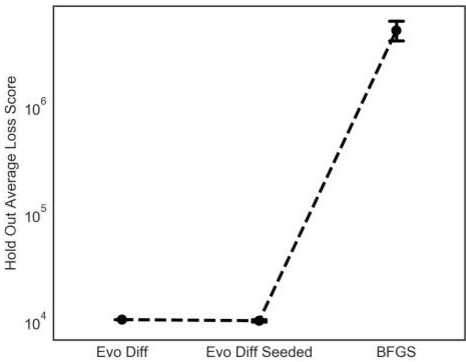
Abstract:The purpose of this research was to compare the robustness and performance of a local and global optimization algorithm when given the task of fitting the parameters of a common non-linear dose-response model utilized in the field of exercise physiology. Traditionally the parameters of dose-response models have been fit using a non-linear least-squares procedure in combination with local optimization algorithms. However, these algorithms have demonstrated limitations in their ability to converge on a globally optimal solution. This research purposes the use of an evolutionary computation based algorithm as an alternative method to fit a nonlinear dose-response model. The results of our comparison over 1000 experimental runs demonstrate the superior performance of the evolutionary computation based algorithm to consistently achieve a stronger model fit and holdout performance in comparison to the local search algorithm. This initial research would suggest that global evolutionary computation based optimization algorithms may present a fast and robust alternative to local algorithms when fitting the parameters of non-linear dose-response models.
High-Resolution Mammogram Synthesis using Progressive Generative Adversarial Networks
Jul 09, 2018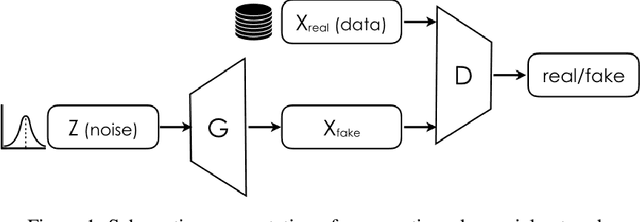
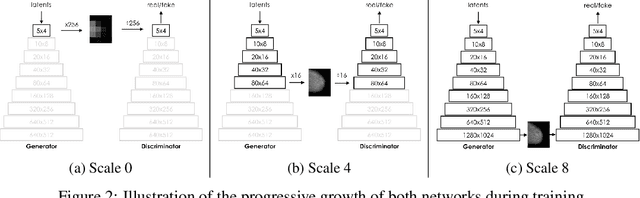
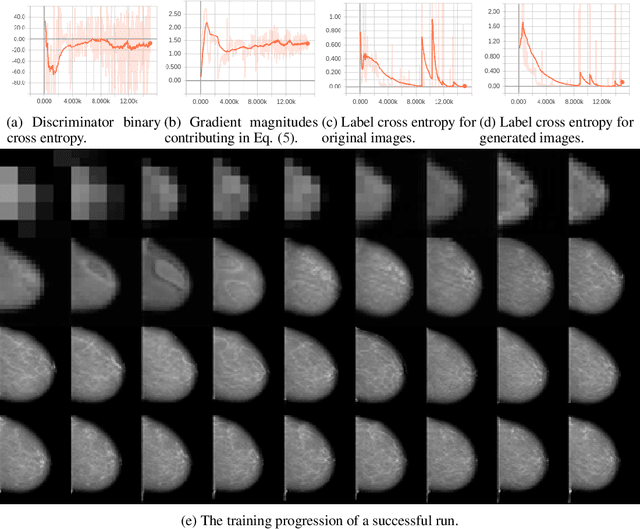
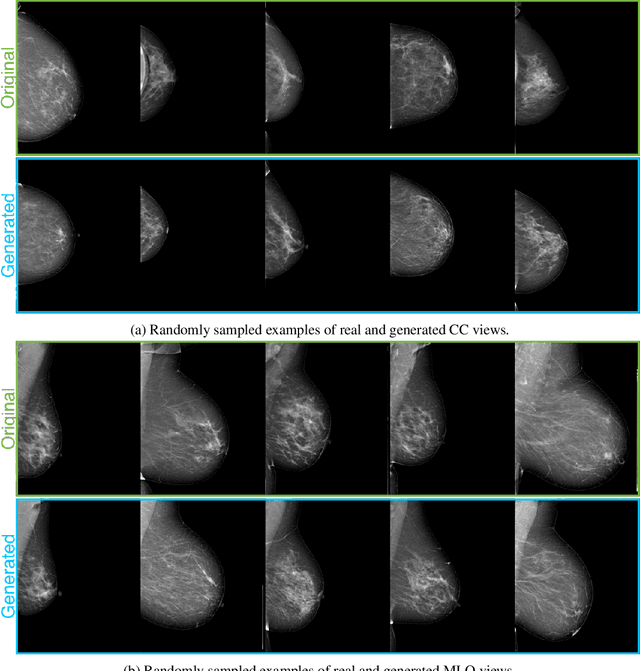
Abstract:The ability to generate synthetic medical images is useful for data augmentation, domain transfer, and out-of-distribution detection. However, generating realistic, high-resolution medical images is challenging, particularly for Full Field Digital Mammograms (FFDM), due to the textural heterogeneity, fine structural details and specific tissue properties. In this paper, we explore the use of progressively trained generative adversarial networks (GANs) to synthesize mammograms, overcoming the underlying instabilities when training such adversarial models. This work is the first to show that generation of realistic synthetic medical images is feasible at up to 1280x1024 pixels, the highest resolution achieved for medical image synthesis, enabling visualizations within standard mammographic hanging protocols. We hope this work can serve as a useful guide and facilitate further research on GANs in the medical imaging domain.
Investigating the Evolvability of Web Page Load Time
Feb 22, 2018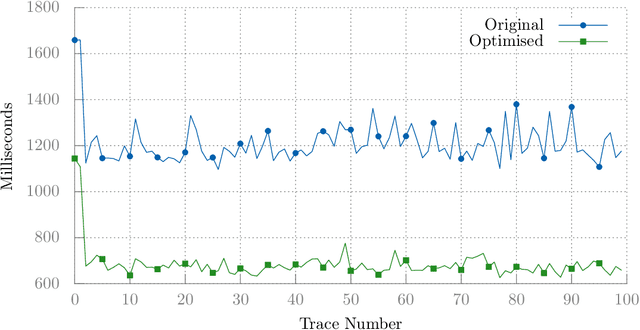
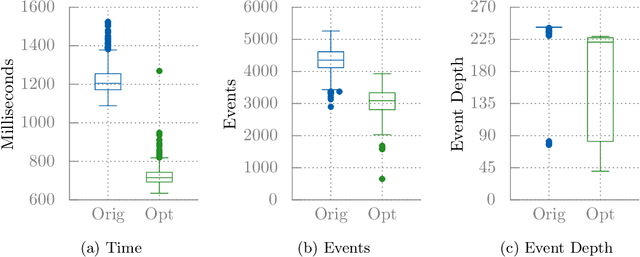
Abstract:Client-side Javascript execution environments (browsers) allow anonymous functions and event-based programming concepts such as callbacks. We investigate whether a mutate-and-test approach can be used to optimise web page load time in these environments. First, we characterise a web page load issue in a benchmark web page and derive performance metrics from page load event traces. We parse Javascript source code to an AST and make changes to method calls which appear in a web page load event trace. We present an operator based solely on code deletion and evaluate an existing "community-contributed" performance optimising code transform. By exploring Javascript code changes and exploiting combinations of non-destructive changes, we can optimise page load time by 41% in our benchmark web page.
PonyGE2: Grammatical Evolution in Python
Apr 26, 2017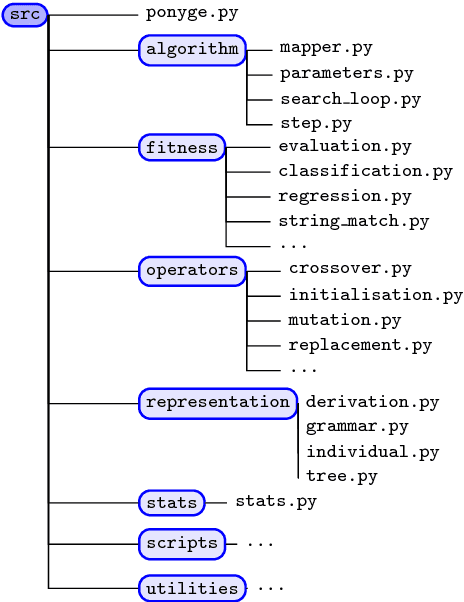
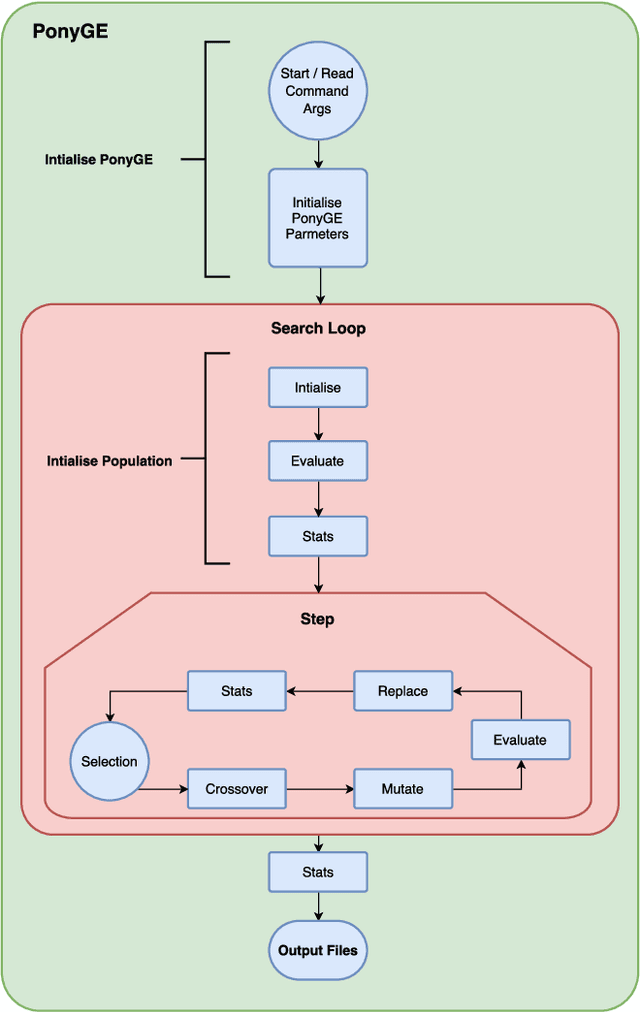
Abstract:Grammatical Evolution (GE) is a population-based evolutionary algorithm, where a formal grammar is used in the genotype to phenotype mapping process. PonyGE2 is an open source implementation of GE in Python, developed at UCD's Natural Computing Research and Applications group. It is intended as an advertisement and a starting-point for those new to GE, a reference for students and researchers, a rapid-prototyping medium for our own experiments, and a Python workout. As well as providing the characteristic genotype to phenotype mapping of GE, a search algorithm engine is also provided. A number of sample problems and tutorials on how to use and adapt PonyGE2 have been developed.
* 8 pages, 4 figures, submitted to the 2017 GECCO Workshop on Evolutionary Computation Software Systems (EvoSoft)
A Search for Improved Performance in Regular Expressions
Apr 13, 2017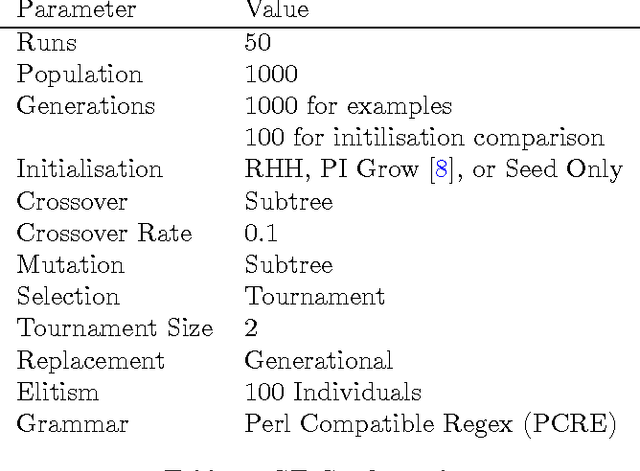
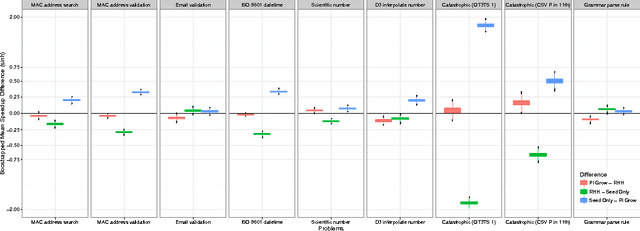

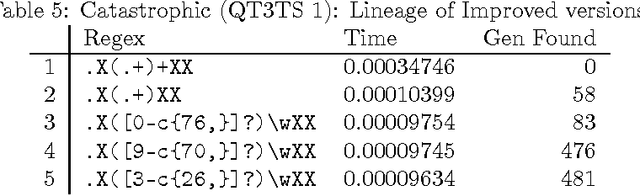
Abstract:The primary aim of automated performance improvement is to reduce the running time of programs while maintaining (or improving on) functionality. In this paper, Genetic Programming is used to find performance improvements in regular expressions for an array of target programs, representing the first application of automated software improvement for run-time performance in the Regular Expression language. This particular problem is interesting as there may be many possible alternative regular expressions which perform the same task while exhibiting subtle differences in performance. A benchmark suite of candidate regular expressions is proposed for improvement. We show that the application of Genetic Programming techniques can result in performance improvements in all cases. As we start evolution from a known good regular expression, diversity is critical in escaping the local optima of the seed expression. In order to understand diversity during evolution we compare an initial population consisting of only seed programs with a population initialised using a combination of a single seed individual with individuals generated using PI Grow and Ramped-half-and-half initialisation mechanisms.
Performance Localisation
Sep 05, 2016



Abstract:Performance becomes an issue particularly when execution cost hinders the functionality of a program. Typically a profiler can be used to find program code execution which represents a large portion of the overall execution cost of a program. Pinpointing where a performance issue exists provides a starting point for tracing cause back through a program. While profiling shows where a performance issue manifests, we use mutation analysis to show where a performance improvement is likely to exist. We find that mutation analysis can indicate locations within a program which are highly impactful to the overall execution cost of a program yet are executed relatively infrequently. By better locating potential performance improvements in programs we hope to make performance improvement more amenable to automation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge