Michael Bockmayr
Multi-Scale Input Strategies for Medulloblastoma Tumor Classification using Deep Transfer Learning
Sep 14, 2021
Abstract:Medulloblastoma (MB) is a primary central nervous system tumor and the most common malignant brain cancer among children. Neuropathologists perform microscopic inspection of histopathological tissue slides under a microscope to assess the severity of the tumor. This is a time-consuming task and often infused with observer variability. Recently, pre-trained convolutional neural networks (CNN) have shown promising results for MB subtype classification. Typically, high-resolution images are divided into smaller tiles for classification, while the size of the tiles has not been systematically evaluated. We study the impact of tile size and input strategy and classify the two major histopathological subtypes-Classic and Demoplastic/Nodular. To this end, we use recently proposed EfficientNets and evaluate tiles with increasing size combined with various downsampling scales. Our results demonstrate using large input tiles pixels followed by intermediate downsampling and patch cropping significantly improves MB classification performance. Our top-performing method achieves the AUC-ROC value of 90.90\% compared to 84.53\% using the previous approach with smaller input tiles.
Medulloblastoma Tumor Classification using Deep Transfer Learning with Multi-Scale EfficientNets
Sep 10, 2021Abstract:Medulloblastoma (MB) is the most common malignant brain tumor in childhood. The diagnosis is generally based on the microscopic evaluation of histopathological tissue slides. However, visual-only assessment of histopathological patterns is a tedious and time-consuming task and is also affected by observer variability. Hence, automated MB tumor classification could assist pathologists by promoting consistency and robust quantification. Recently, convolutional neural networks (CNNs) have been proposed for this task, while transfer learning has shown promising results. In this work, we propose an end-to-end MB tumor classification and explore transfer learning with various input sizes and matching network dimensions. We focus on differentiating between the histological subtypes classic and desmoplastic/nodular. For this purpose, we systematically evaluate recently proposed EfficientNets, which uniformly scale all dimensions of a CNN. Using a data set with 161 cases, we demonstrate that pre-trained EfficientNets with larger input resolutions lead to significant performance improvements compared to commonly used pre-trained CNN architectures. Also, we highlight the importance of transfer learning, when using such large architectures. Overall, our best performing method achieves an F1-Score of 80.1%.
Resolving challenges in deep learning-based analyses of histopathological images using explanation methods
Aug 15, 2019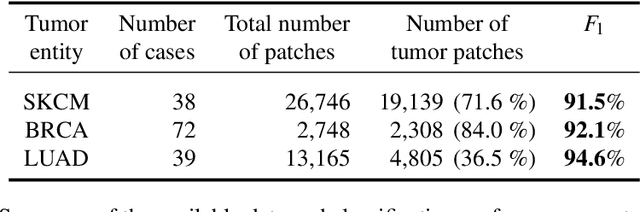
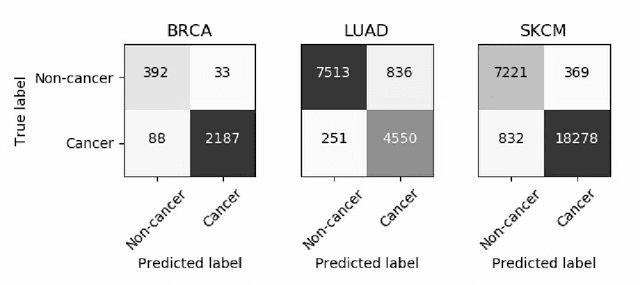
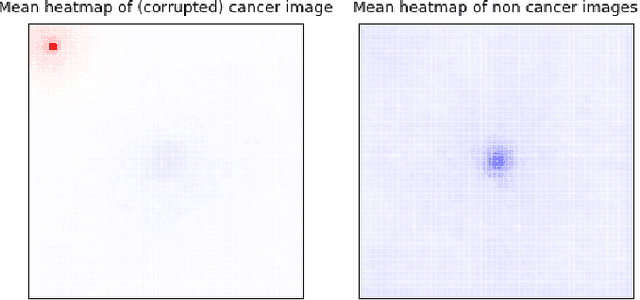
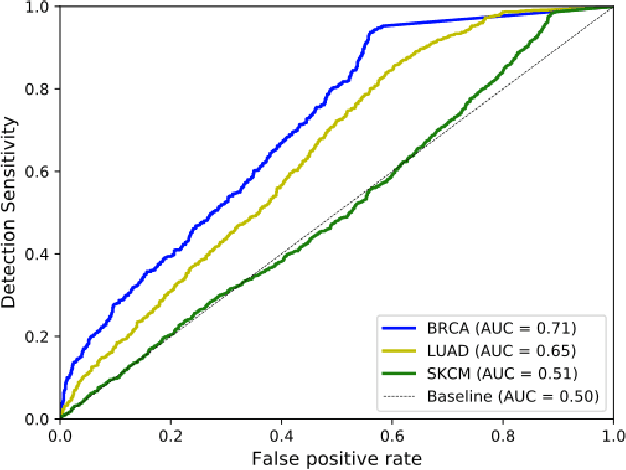
Abstract:Deep learning has recently gained popularity in digital pathology due to its high prediction quality. However, the medical domain requires explanation and insight for a better understanding beyond standard quantitative performance evaluation. Recently, explanation methods have emerged, which are so far still rarely used in medicine. This work shows their application to generate heatmaps that allow to resolve common challenges encountered in deep learning-based digital histopathology analyses. These challenges comprise biases typically inherent to histopathology data. We study binary classification tasks of tumor tissue discrimination in publicly available haematoxylin and eosin slides of various tumor entities and investigate three types of biases: (1) biases which affect the entire dataset, (2) biases which are by chance correlated with class labels and (3) sampling biases. While standard analyses focus on patch-level evaluation, we advocate pixel-wise heatmaps, which offer a more precise and versatile diagnostic instrument and furthermore help to reveal biases in the data. This insight is shown to not only detect but also to be helpful to remove the effects of common hidden biases, which improves generalization within and across datasets. For example, we could see a trend of improved area under the receiver operating characteristic curve by 5% when reducing a labeling bias. Explanation techniques are thus demonstrated to be a helpful and highly relevant tool for the development and the deployment phases within the life cycle of real-world applications in digital pathology.
Towards computational fluorescence microscopy: Machine learning-based integrated prediction of morphological and molecular tumor profiles
May 28, 2018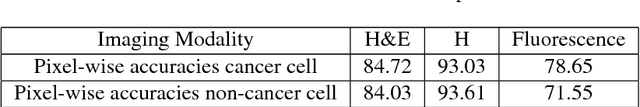


Abstract:Recent advances in cancer research largely rely on new developments in microscopic or molecular profiling techniques offering high level of detail with respect to either spatial or molecular features, but usually not both. Here, we present a novel machine learning-based computational approach that allows for the identification of morphological tissue features and the prediction of molecular properties from breast cancer imaging data. This integration of microanatomic information of tumors with complex molecular profiling data, including protein or gene expression, copy number variation, gene methylation and somatic mutations, provides a novel means to computationally score molecular markers with respect to their relevance to cancer and their spatial associations within the tumor microenvironment.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge