Resolving challenges in deep learning-based analyses of histopathological images using explanation methods
Paper and Code
Aug 15, 2019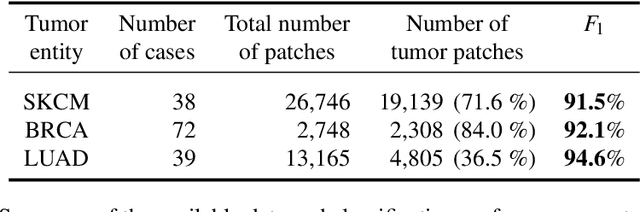
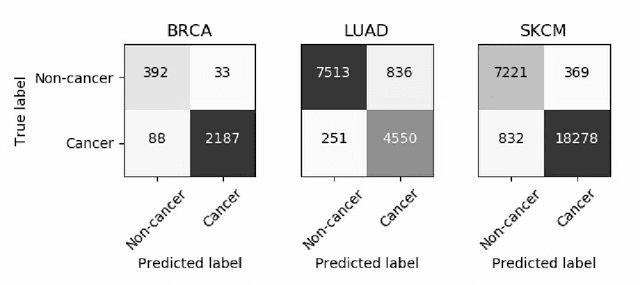
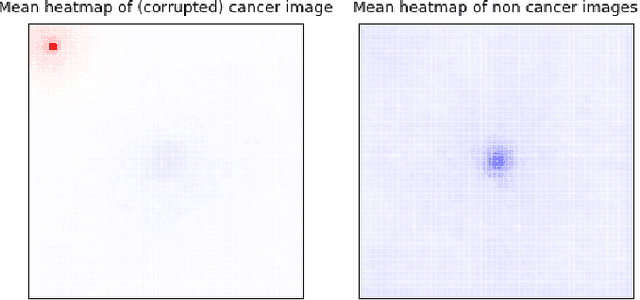
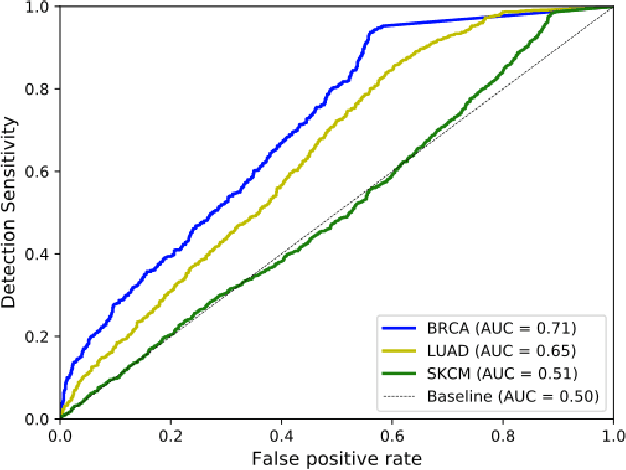
Deep learning has recently gained popularity in digital pathology due to its high prediction quality. However, the medical domain requires explanation and insight for a better understanding beyond standard quantitative performance evaluation. Recently, explanation methods have emerged, which are so far still rarely used in medicine. This work shows their application to generate heatmaps that allow to resolve common challenges encountered in deep learning-based digital histopathology analyses. These challenges comprise biases typically inherent to histopathology data. We study binary classification tasks of tumor tissue discrimination in publicly available haematoxylin and eosin slides of various tumor entities and investigate three types of biases: (1) biases which affect the entire dataset, (2) biases which are by chance correlated with class labels and (3) sampling biases. While standard analyses focus on patch-level evaluation, we advocate pixel-wise heatmaps, which offer a more precise and versatile diagnostic instrument and furthermore help to reveal biases in the data. This insight is shown to not only detect but also to be helpful to remove the effects of common hidden biases, which improves generalization within and across datasets. For example, we could see a trend of improved area under the receiver operating characteristic curve by 5% when reducing a labeling bias. Explanation techniques are thus demonstrated to be a helpful and highly relevant tool for the development and the deployment phases within the life cycle of real-world applications in digital pathology.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge