Mehdi Amian
QU-BraTS: MICCAI BraTS 2020 Challenge on Quantifying Uncertainty in Brain Tumor Segmentation -- Analysis of Ranking Metrics and Benchmarking Results
Dec 19, 2021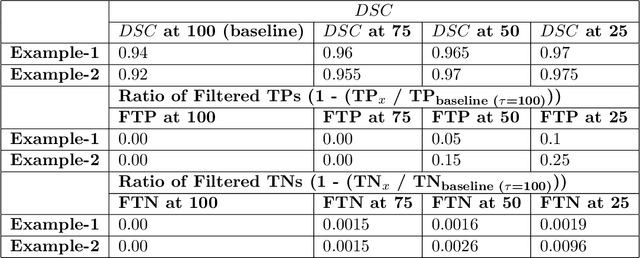
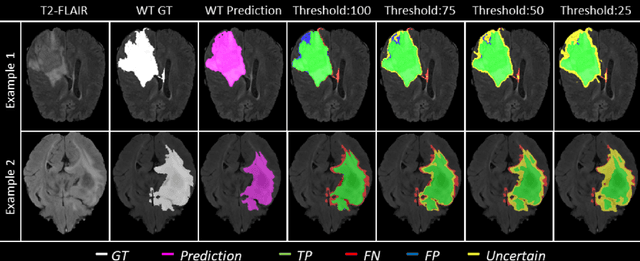
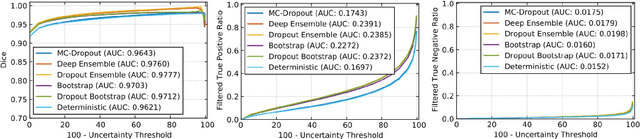
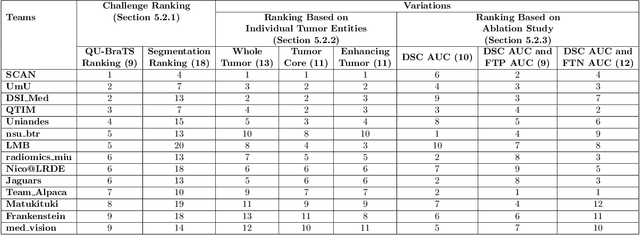
Abstract:Deep learning (DL) models have provided the state-of-the-art performance in a wide variety of medical imaging benchmarking challenges, including the Brain Tumor Segmentation (BraTS) challenges. However, the task of focal pathology multi-compartment segmentation (e.g., tumor and lesion sub-regions) is particularly challenging, and potential errors hinder the translation of DL models into clinical workflows. Quantifying the reliability of DL model predictions in the form of uncertainties, could enable clinical review of the most uncertain regions, thereby building trust and paving the way towards clinical translation. Recently, a number of uncertainty estimation methods have been introduced for DL medical image segmentation tasks. Developing metrics to evaluate and compare the performance of uncertainty measures will assist the end-user in making more informed decisions. In this study, we explore and evaluate a metric developed during the BraTS 2019-2020 task on uncertainty quantification (QU-BraTS), and designed to assess and rank uncertainty estimates for brain tumor multi-compartment segmentation. This metric (1) rewards uncertainty estimates that produce high confidence in correct assertions, and those that assign low confidence levels at incorrect assertions, and (2) penalizes uncertainty measures that lead to a higher percentages of under-confident correct assertions. We further benchmark the segmentation uncertainties generated by 14 independent participating teams of QU-BraTS 2020, all of which also participated in the main BraTS segmentation task. Overall, our findings confirm the importance and complementary value that uncertainty estimates provide to segmentation algorithms, and hence highlight the need for uncertainty quantification in medical image analyses. Our evaluation code is made publicly available at https://github.com/RagMeh11/QU-BraTS.
Improving the Algorithm of Deep Learning with Differential Privacy
Jul 12, 2021
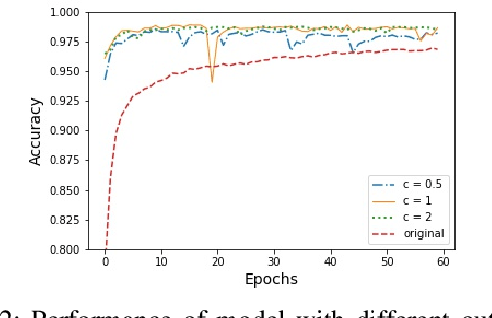
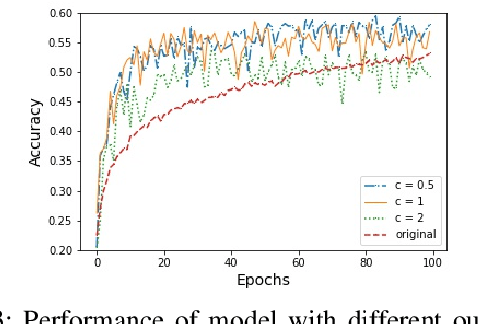
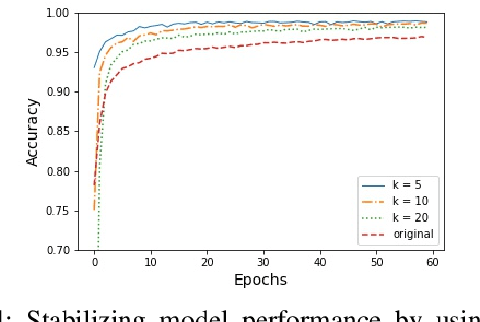
Abstract:In this paper, an adjustment to the original differentially private stochastic gradient descent (DPSGD) algorithm for deep learning models is proposed. As a matter of motivation, to date, almost no state-of-the-art machine learning algorithm hires the existing privacy protecting components due to otherwise serious compromise in their utility despite the vital necessity. The idea in this study is natural and interpretable, contributing to improve the utility with respect to the state-of-the-art. Another property of the proposed technique is its simplicity which makes it again more natural and also more appropriate for real world and specially commercial applications. The intuition is to trim and balance out wild individual discrepancies for privacy reasons, and at the same time, to preserve relative individual differences for seeking performance. The idea proposed here can also be applied to the recurrent neural networks (RNN) to solve the gradient exploding problem. The algorithm is applied to benchmark datasets MNIST and CIFAR-10 for a classification task and the utility measure is calculated. The results outperformed the original work.
Multi-Resolution 3D CNN for MRI Brain Tumor Segmentation and Survival Prediction
Nov 19, 2019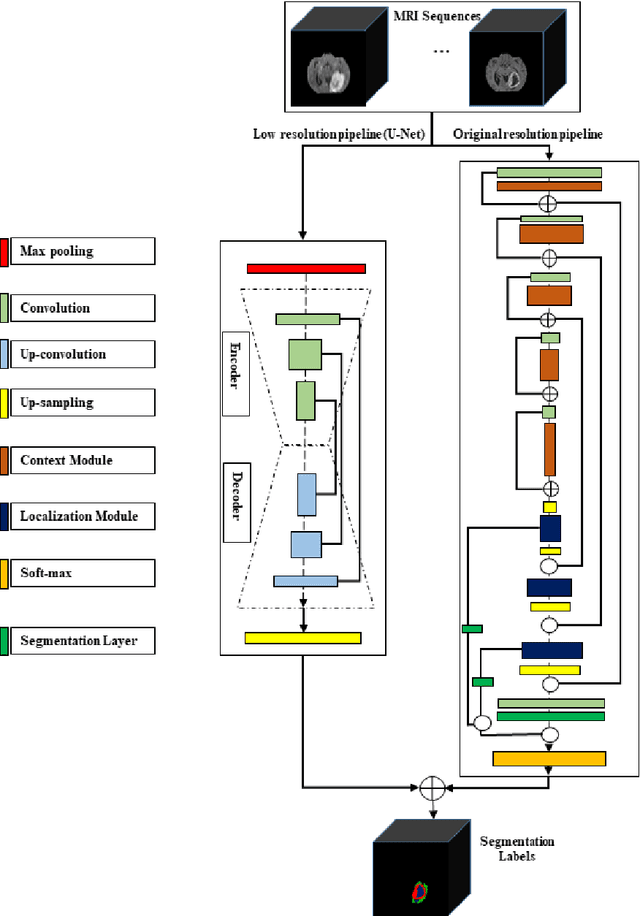
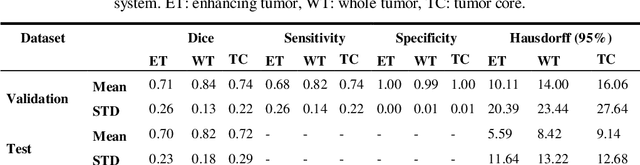
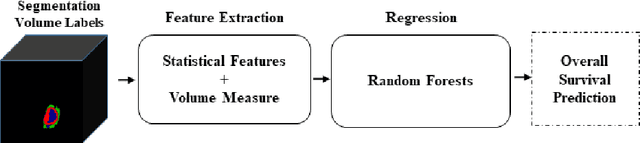

Abstract:In this study, an automated three dimensional (3D) deep segmentation approach for detecting gliomas in 3D pre-operative MRI scans is proposed. Then, a classi-fication algorithm based on random forests, for survival prediction is presented. The objective is to segment the glioma area and produce segmentation labels for its different sub-regions, i.e. necrotic and the non-enhancing tumor core, the peri-tumoral edema, and enhancing tumor. The proposed deep architecture for the segmentation task encompasses two parallel streamlines with two different reso-lutions. One deep convolutional neural network is to learn local features of the input data while the other one is set to have a global observation on whole image. Deemed to be complementary, the outputs of each stream are then merged to pro-vide an ensemble complete learning of the input image. The proposed network takes the whole image as input instead of patch-based approaches in order to con-sider the semantic features throughout the whole volume. The algorithm is trained on BraTS 2019 which included 335 training cases, and validated on 127 unseen cases from the validation dataset using a blind testing approach. The proposed method was also evaluated on the BraTS 2019 challenge test dataset of 166 cases. The results show that the proposed methods provide promising segmentations as well as survival prediction. The mean Dice overlap measures of automatic brain tumor segmentation for validation set were 0.84, 0.74 and 0.71 for the whole tu-mor, core and enhancing tumor, respectively. The corresponding results for the challenge test dataset were 0.82, 0.72, and 0.70, respectively. The overall accura-cy of the proposed model for the survival prediction task is %52 for the valida-tion and %49 for the test dataset.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge