Martin Rosvall
Insights from Network Science can advance Deep Graph Learning
Feb 03, 2025Abstract:Deep graph learning and network science both analyze graphs but approach similar problems from different perspectives. Whereas network science focuses on models and measures that reveal the organizational principles of complex systems with explicit assumptions, deep graph learning focuses on flexible and generalizable models that learn patterns in graph data in an automated fashion. Despite these differences, both fields share the same goal: to better model and understand patterns in graph-structured data. Early efforts to integrate methods, models, and measures from network science and deep graph learning indicate significant untapped potential. In this position, we explore opportunities at their intersection. We discuss open challenges in deep graph learning, including data augmentation, improved evaluation practices, higher-order models, and pooling methods. Likewise, we highlight challenges in network science, including scaling to massive graphs, integrating continuous gradient-based optimization, and developing standardized benchmarks.
A Correlation- and Mean-Aware Loss Function and Benchmarking Framework to Improve GAN-based Tabular Data Synthesis
May 27, 2024Abstract:Advancements in science rely on data sharing. In medicine, where personal data are often involved, synthetic tabular data generated by generative adversarial networks (GANs) offer a promising avenue. However, existing GANs struggle to capture the complexities of real-world tabular data, which often contain a mix of continuous and categorical variables with potential imbalances and dependencies. We propose a novel correlation- and mean-aware loss function designed to address these challenges as a regularizer for GANs. To ensure a rigorous evaluation, we establish a comprehensive benchmarking framework using ten real-world datasets and eight established tabular GAN baselines. The proposed loss function demonstrates statistically significant improvements over existing methods in capturing the true data distribution, significantly enhancing the quality of synthetic data generated with GANs. The benchmarking framework shows that the enhanced synthetic data quality leads to improved performance in downstream machine learning (ML) tasks, ultimately paving the way for easier data sharing.
Module-based regularization improves Gaussian graphical models when observing noisy data
Apr 04, 2023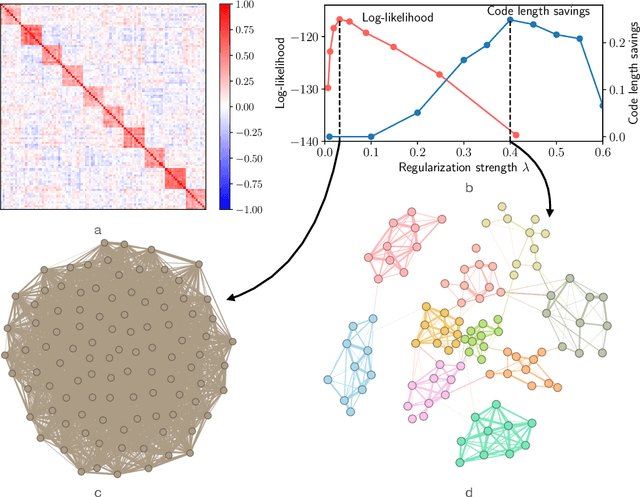
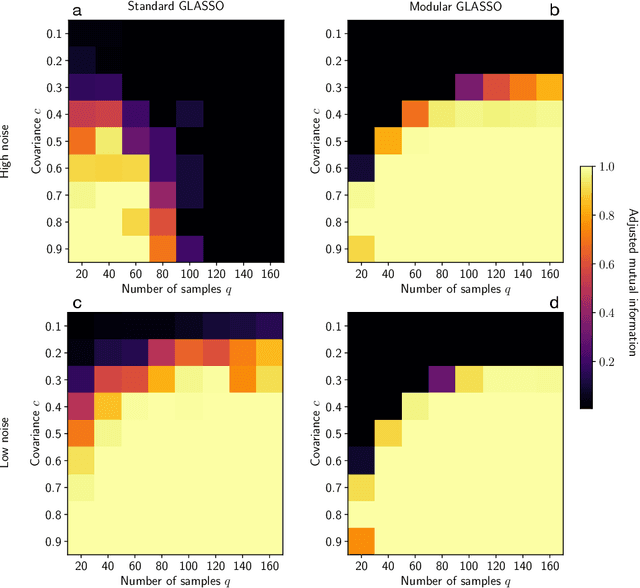
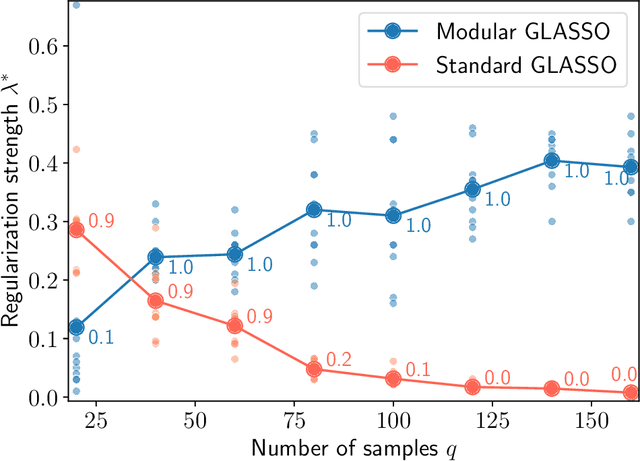
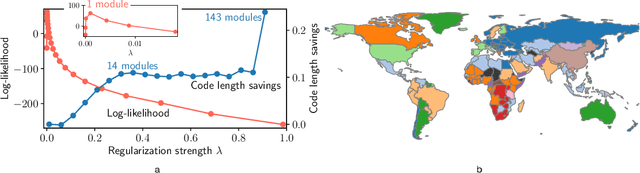
Abstract:Researchers often represent relations in multi-variate correlational data using Gaussian graphical models, which require regularization to sparsify the models. Acknowledging that they often study the modular structure of the inferred network, we suggest integrating it in the cross-validation of the regularization strength to balance under- and overfitting. Using synthetic and real data, we show that this approach allows us to better recover and infer modular structure in noisy data compared with the graphical lasso, a standard approach using the Gaussian log-likelihood when cross-validating the regularization strength.
Mapping higher-order network flows in memory and multilayer networks with Infomap
Oct 16, 2017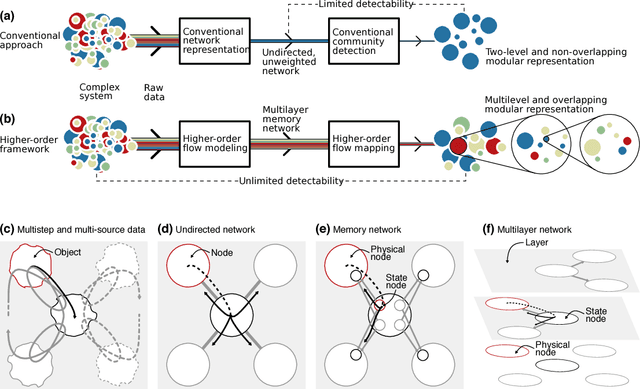
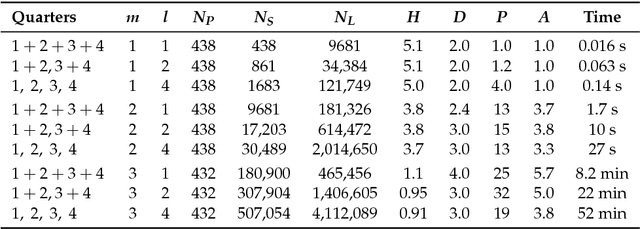
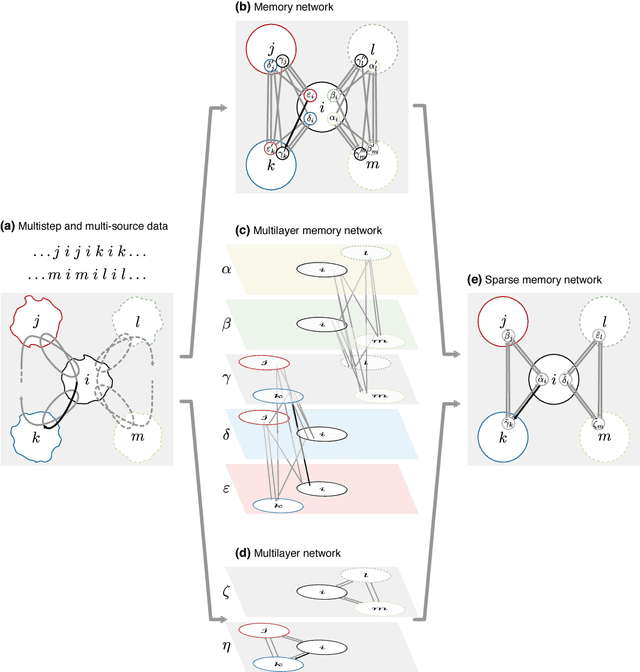
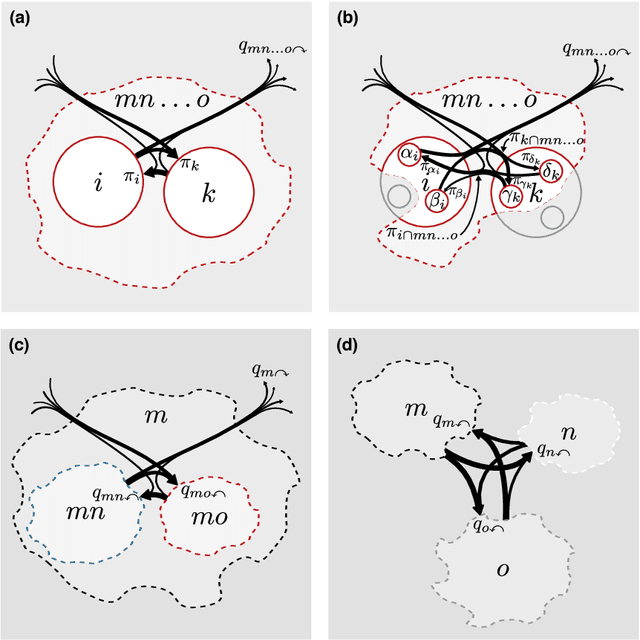
Abstract:Comprehending complex systems by simplifying and highlighting important dynamical patterns requires modeling and mapping higher-order network flows. However, complex systems come in many forms and demand a range of representations, including memory and multilayer networks, which in turn call for versatile community-detection algorithms to reveal important modular regularities in the flows. Here we show that various forms of higher-order network flows can be represented in a unified way with networks that distinguish physical nodes for representing a~complex system's objects from state nodes for describing flows between the objects. Moreover, these so-called sparse memory networks allow the information-theoretic community detection method known as the map equation to identify overlapping and nested flow modules in data from a range of~different higher-order interactions such as multistep, multi-source, and temporal data. We derive the map equation applied to sparse memory networks and describe its search algorithm Infomap, which can exploit the flexibility of sparse memory networks. Together they provide a general solution to reveal overlapping modular patterns in higher-order flows through complex systems.
* 23 pages, 4 figures
Modeling sequences and temporal networks with dynamic community structures
Sep 20, 2017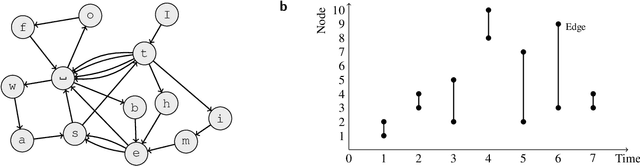
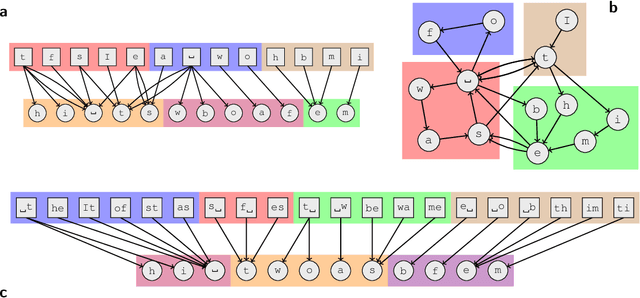
Abstract:In evolving complex systems such as air traffic and social organizations, collective effects emerge from their many components' dynamic interactions. While the dynamic interactions can be represented by temporal networks with nodes and links that change over time, they remain highly complex. It is therefore often necessary to use methods that extract the temporal networks' large-scale dynamic community structure. However, such methods are subject to overfitting or suffer from effects of arbitrary, a priori imposed timescales, which should instead be extracted from data. Here we simultaneously address both problems and develop a principled data-driven method that determines relevant timescales and identifies patterns of dynamics that take place on networks as well as shape the networks themselves. We base our method on an arbitrary-order Markov chain model with community structure, and develop a nonparametric Bayesian inference framework that identifies the simplest such model that can explain temporal interaction data.
* 15 Pages, 6 figures, 2 tables
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge