Martin Bøgsted
On the Relation Between Autoencoders and Non-negative Matrix Factorization, and Their Application for Mutational Signature Extraction
May 13, 2024Abstract:The aim of this study is to provide a foundation to understand the relationship between non-negative matrix factorization (NMF) and non-negative autoencoders enabling proper interpretation and understanding of autoencoder-based alternatives to NMF. Since its introduction, NMF has been a popular tool for extracting interpretable, low-dimensional representations of high-dimensional data. However, recently, several studies have proposed to replace NMF with autoencoders. This increasing popularity of autoencoders warrants an investigation on whether this replacement is in general valid and reasonable. Moreover, the exact relationship between non-negative autoencoders and NMF has not been thoroughly explored. Thus, a main aim of this study is to investigate in detail the relationship between non-negative autoencoders and NMF. We find that the connection between the two models can be established through convex NMF, which is a restricted case of NMF. In particular, convex NMF is a special case of an autoencoder. The performance of NMF and autoencoders is compared within the context of extraction of mutational signatures from cancer genomics data. We find that the reconstructions based on NMF are more accurate compared to autoencoders, while the signatures extracted using both methods show comparable consistencies and values when externally validated. These findings suggest that the non-negative autoencoders investigated in this article do not provide an improvement of NMF in the field of mutational signature extraction.
A primer on synthetic health data
Jan 31, 2024Abstract:Recent advances in deep generative models have greatly expanded the potential to create realistic synthetic health datasets. These synthetic datasets aim to preserve the characteristics, patterns, and overall scientific conclusions derived from sensitive health datasets without disclosing patient identity or sensitive information. Thus, synthetic data can facilitate safe data sharing that supports a range of initiatives including the development of new predictive models, advanced health IT platforms, and general project ideation and hypothesis development. However, many questions and challenges remain, including how to consistently evaluate a synthetic dataset's similarity and predictive utility in comparison to the original real dataset and risk to privacy when shared. Additional regulatory and governance issues have not been widely addressed. In this primer, we map the state of synthetic health data, including generation and evaluation methods and tools, existing examples of deployment, the regulatory and ethical landscape, access and governance options, and opportunities for further development.
Regression on imperfect class labels derived by unsupervised clustering
Aug 16, 2019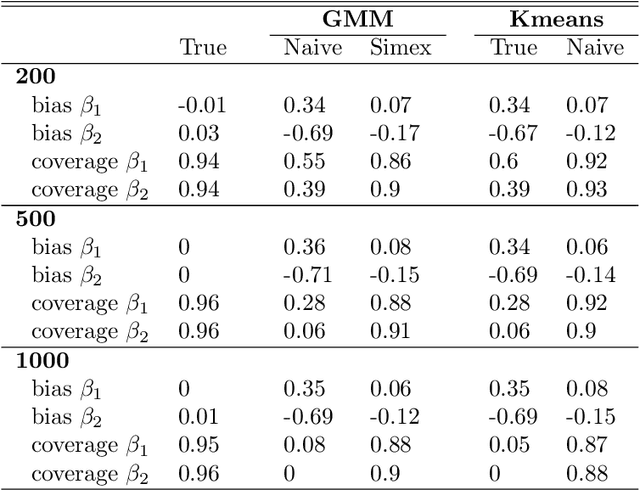
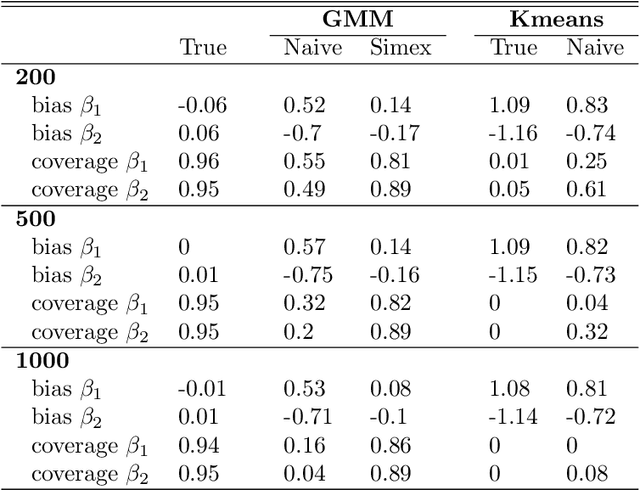
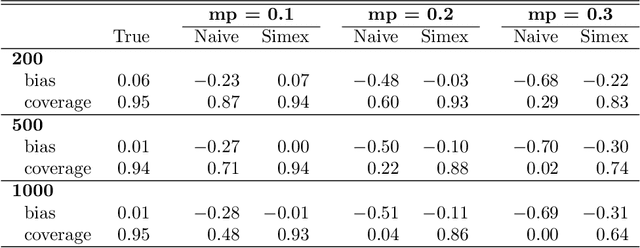
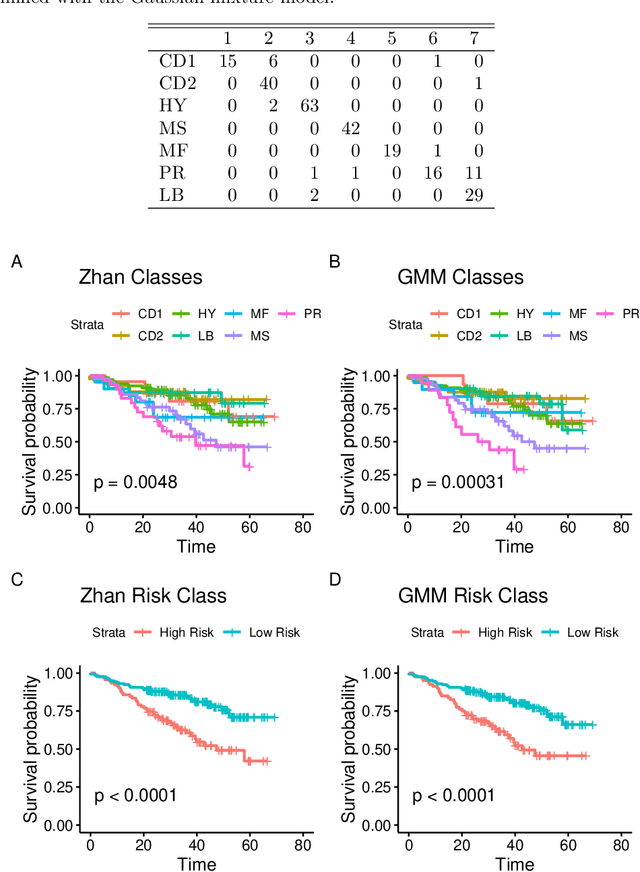
Abstract:Outcome regressed on class labels identified by unsupervised clustering is custom in many applications. However, it is common to ignore the misclassification of class labels caused by the learning algorithm, which potentially leads to serious bias of the estimated effect parameters. Due to its generality we suggest to redress the situation by use of the simulation and extrapolation method. Performance is illustrated by simulated data from Gaussian mixture models. Finally, we apply our method to a study which regressed overall survival on class labels derived from unsupervised clustering of gene expression data from bone marrow samples of multiple myeloma patients.
Estimating a common covariance matrix for network meta-analysis of gene expression datasets in diffuse large B-cell lymphoma
Aug 21, 2017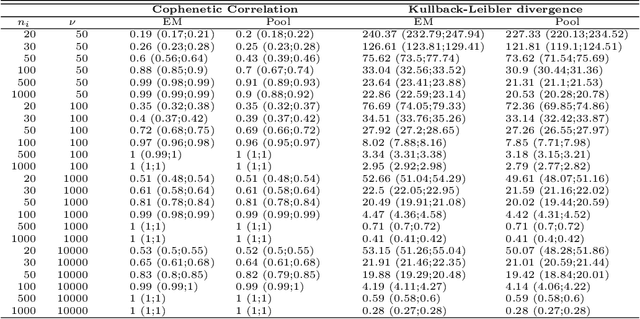

Abstract:The estimation of covariance matrices of gene expressions has many applications in cancer systems biology. Many gene expression studies, however, are hampered by low sample size and it has therefore become popular to increase sample size by collecting gene expression data across studies. Motivated by the traditional meta-analysis using random effects models, we present a hierarchical random covariance model and use it for the meta-analysis of gene correlation networks across 11 large-scale gene expression studies of diffuse large B-cell lymphoma (DLBCL). We suggest to use a maximum likelihood estimator for the underlying common covariance matrix and introduce an EM algorithm for estimation. By simulation experiments comparing the estimated covariance matrices by cophenetic correlation and Kullback-Leibler divergence the suggested estimator showed to perform better or not worse than a simple pooled estimator. In a posthoc analysis of the estimated common covariance matrix for the DLBCL data we were able to identify novel biologically meaningful gene correlation networks with eigengenes of prognostic value. In conclusion, the method seems to provide a generally applicable framework for meta-analysis, when multiple features are measured and believed to share a common covariance matrix obscured by study dependent noise.
Targeted Fused Ridge Estimation of Inverse Covariance Matrices from Multiple High-Dimensional Data Classes
Sep 26, 2015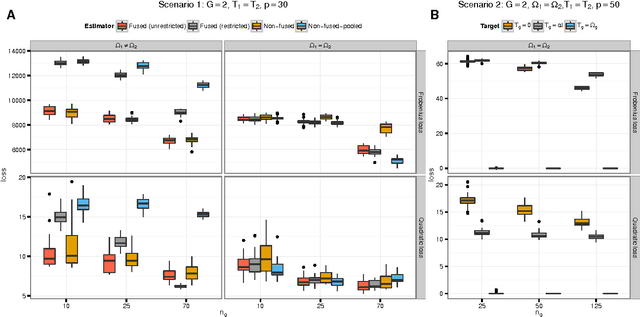
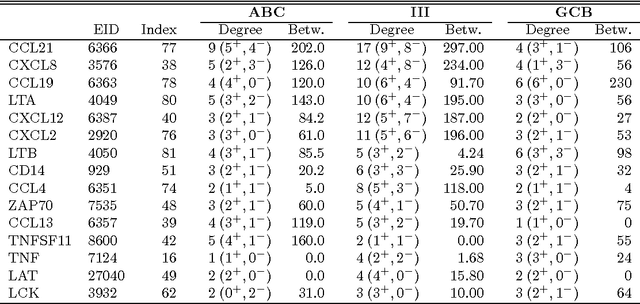
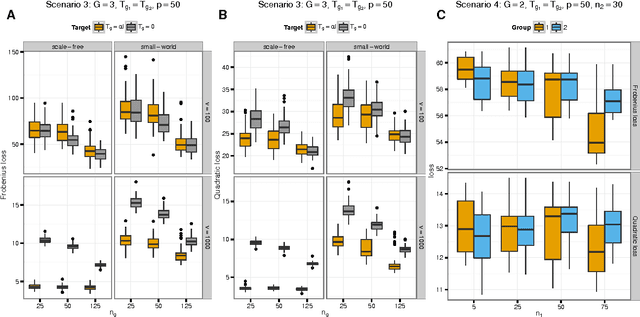
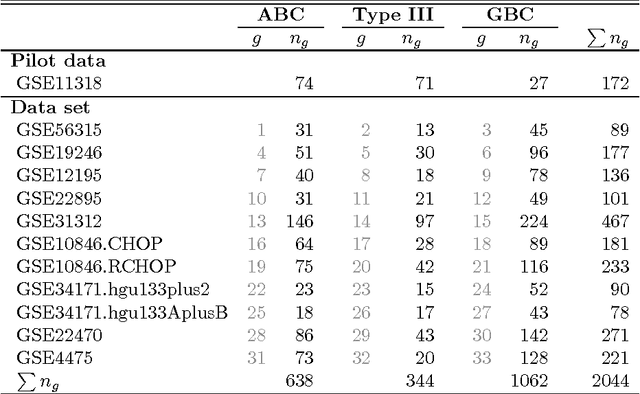
Abstract:We consider the problem of jointly estimating multiple precision matrices from (aggregated) high-dimensional data consisting of distinct classes. An $\ell_2$-penalized maximum-likelihood approach is employed. The suggested approach is flexible and generic, incorporating several other $\ell_2$-penalized estimators as special cases. In addition, the approach allows for the specification of target matrices through which prior knowledge may be incorporated and which can stabilize the estimation procedure in high-dimensional settings. The result is a targeted fused ridge estimator that is of use when the precision matrices of the constituent classes are believed to chiefly share the same structure while potentially differing in a number of locations of interest. It has many applications in (multi)factorial study designs. We focus on the graphical interpretation of precision matrices with the proposed estimator then serving as a basis for integrative or meta-analytic Gaussian graphical modeling. Situations are considered in which the classes are defined by data sets and/or (subtypes of) diseases. The performance of the proposed estimator in the graphical modeling setting is assessed through extensive simulation experiments. Its practical usability is illustrated by the differential network modeling of 11 large-scale diffuse large B-cell lymphoma gene expression data sets. The estimator and its related procedures are incorporated into the R-package rags2ridges.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge