Marta Cremonesi
Application of the nnU-Net for automatic segmentation of lung lesion on CT images, and implication on radiomic models
Sep 24, 2022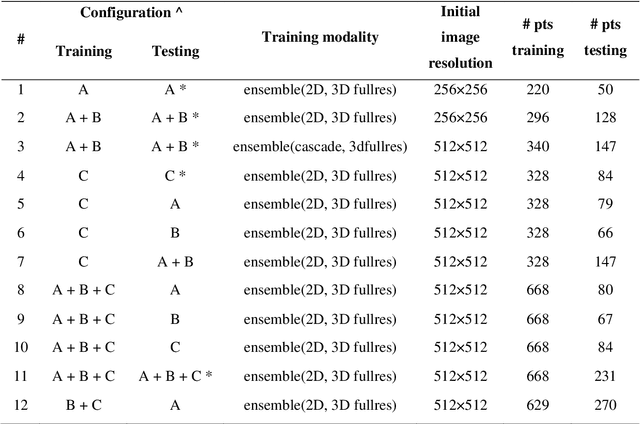
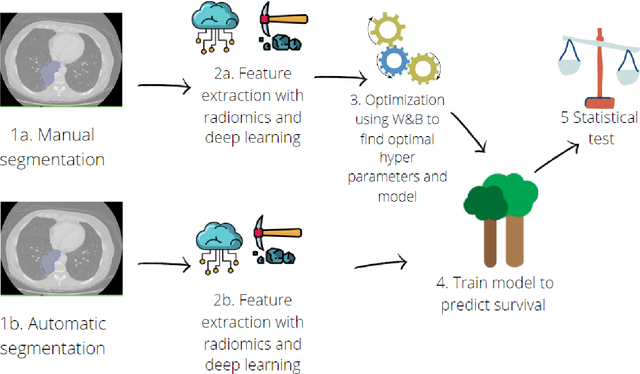
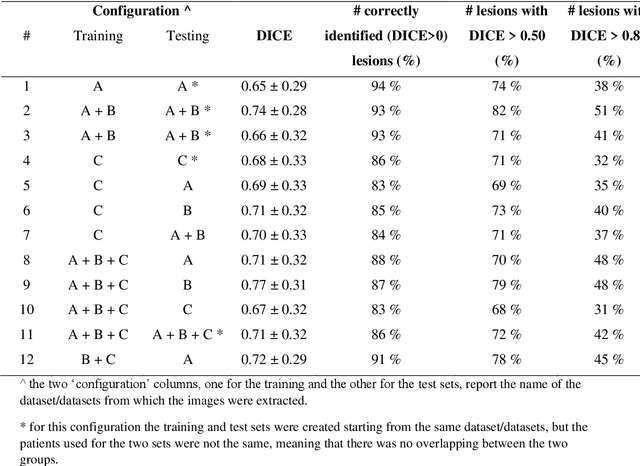
Abstract:Lesion segmentation is a crucial step of the radiomic workflow. Manual segmentation requires long execution time and is prone to variability, impairing the realisation of radiomic studies and their robustness. In this study, a deep-learning automatic segmentation method was applied on computed tomography images of non-small-cell lung cancer patients. The use of manual vs automatic segmentation in the performance of survival radiomic models was assessed, as well. METHODS A total of 899 NSCLC patients were included (2 proprietary: A and B, 1 public datasets: C). Automatic segmentation of lung lesions was performed by training a previously developed architecture, the nnU-Net, including 2D, 3D and cascade approaches. The quality of automatic segmentation was evaluated with DICE coefficient, considering manual contours as reference. The impact of automatic segmentation on the performance of a radiomic model for patient survival was explored by extracting radiomic hand-crafted and deep-learning features from manual and automatic contours of dataset A, and feeding different machine learning algorithms to classify survival above/below median. Models' accuracies were assessed and compared. RESULTS The best agreement between automatic and manual contours with DICE=0.78 +(0.12) was achieved by averaging predictions from 2D and 3D models, and applying a post-processing technique to extract the maximum connected component. No statistical differences were observed in the performances of survival models when using manual or automatic contours, hand-crafted, or deep features. The best classifier showed an accuracy between 0.65 and 0.78. CONCLUSION The promising role of nnU-Net for automatic segmentation of lung lesions was confirmed, dramatically reducing the time-consuming physicians' workload without impairing the accuracy of survival predictive models based on radiomics.
A multicenter study on radiomic features from T$_2$-weighted images of a customized MR pelvic phantom setting the basis for robust radiomic models in clinics
May 18, 2020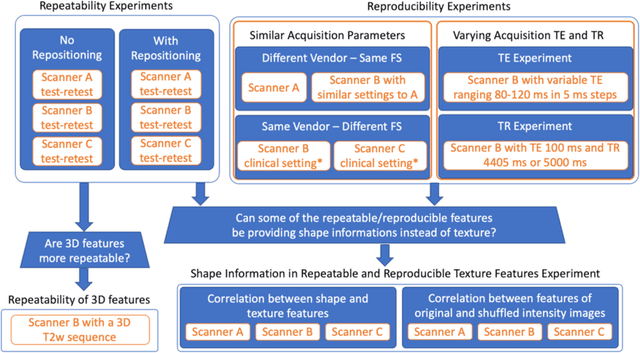
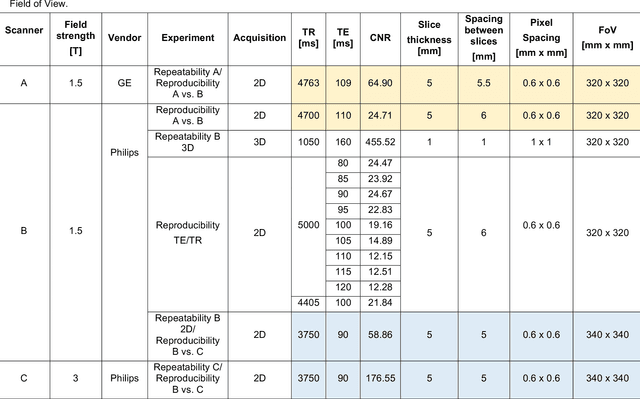
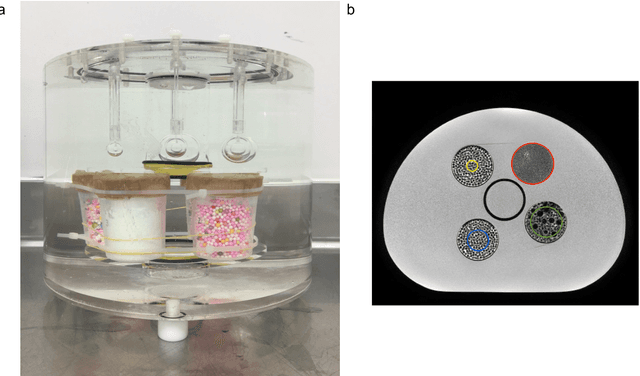
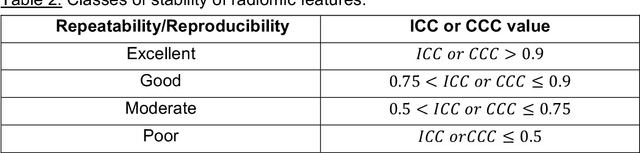
Abstract:In this study we investigated the repeatability and reproducibility of radiomic features extracted from MRI images and provide a workflow to identify robust features. 2D and 3D T$_2$-weighted images of a pelvic phantom were acquired on three scanners of two manufacturers and two magnetic field strengths. The repeatability and reproducibility of the radiomic features were assessed respectively by intraclass correlation coefficient (ICC) and concordance correlation coefficient (CCC), considering repeated acquisitions with or without phantom repositioning, and with different scanner/acquisition type, and acquisition parameters. The features showing ICC/CCC > 0.9 were selected, and their dependence on shape information (Spearman's $\rho$> 0.8) was analyzed. They were classified for their ability to distinguish textures, after shuffling voxel intensities. From 944 2D features, 79.9% to 96.4% showed excellent repeatability in fixed position across all scanners. Much lower range (11.2% to 85.4%) was obtained after phantom repositioning. 3D extraction did not improve repeatability performance. Excellent reproducibility between scanners was observed in 4.6% to 15.6% of the features, at fixed imaging parameters. 82.4% to 94.9% of features showed excellent agreement when extracted from images acquired with TEs 5 ms apart (values decreased when increasing TE intervals) and 90.7% of the features exhibited excellent reproducibility for changes in TR. 2.0% of non-shape features were identified as providing only shape information. This study demonstrates that radiomic features are affected by specific MRI protocols. The use of our radiomic pelvic phantom allowed to identify unreliable features for radiomic analysis on T$_2$-weighted images. This paper proposes a general workflow to identify repeatable, reproducible, and informative radiomic features, fundamental to ensure robustness of clinical studies.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge