Nuno Loução
Counterfactual MRI Data Augmentation using Conditional Denoising Diffusion Generative Models
Oct 31, 2024



Abstract:Deep learning (DL) models in medical imaging face challenges in generalizability and robustness due to variations in image acquisition parameters (IAP). In this work, we introduce a novel method using conditional denoising diffusion generative models (cDDGMs) to generate counterfactual magnetic resonance (MR) images that simulate different IAP without altering patient anatomy. We demonstrate that using these counterfactual images for data augmentation can improve segmentation accuracy, particularly in out-of-distribution settings, enhancing the overall generalizability and robustness of DL models across diverse imaging conditions. Our approach shows promise in addressing domain and covariate shifts in medical imaging. The code is publicly available at https: //github.com/pedromorao/Counterfactual-MRI-Data-Augmentation
A multicenter study on radiomic features from T$_2$-weighted images of a customized MR pelvic phantom setting the basis for robust radiomic models in clinics
May 18, 2020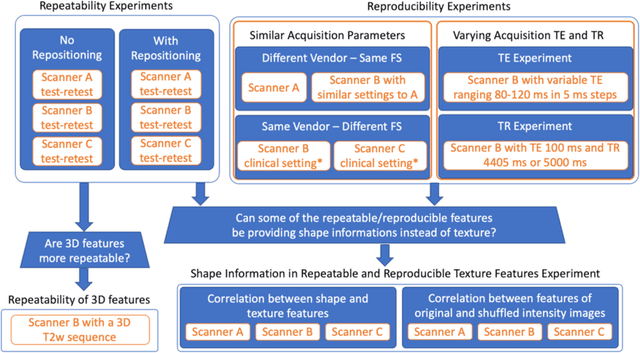
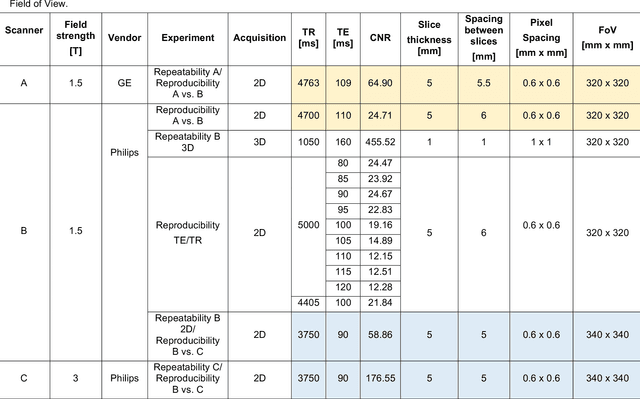
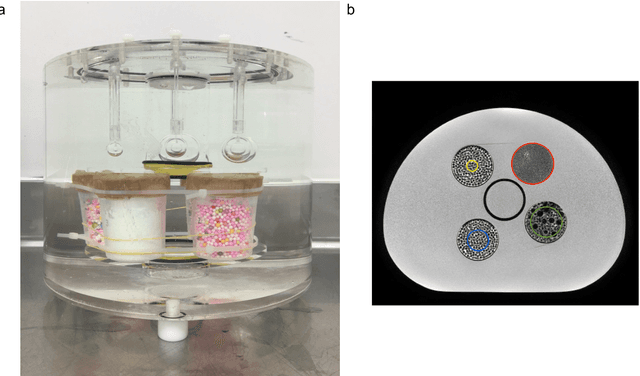
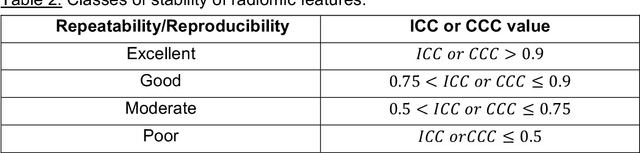
Abstract:In this study we investigated the repeatability and reproducibility of radiomic features extracted from MRI images and provide a workflow to identify robust features. 2D and 3D T$_2$-weighted images of a pelvic phantom were acquired on three scanners of two manufacturers and two magnetic field strengths. The repeatability and reproducibility of the radiomic features were assessed respectively by intraclass correlation coefficient (ICC) and concordance correlation coefficient (CCC), considering repeated acquisitions with or without phantom repositioning, and with different scanner/acquisition type, and acquisition parameters. The features showing ICC/CCC > 0.9 were selected, and their dependence on shape information (Spearman's $\rho$> 0.8) was analyzed. They were classified for their ability to distinguish textures, after shuffling voxel intensities. From 944 2D features, 79.9% to 96.4% showed excellent repeatability in fixed position across all scanners. Much lower range (11.2% to 85.4%) was obtained after phantom repositioning. 3D extraction did not improve repeatability performance. Excellent reproducibility between scanners was observed in 4.6% to 15.6% of the features, at fixed imaging parameters. 82.4% to 94.9% of features showed excellent agreement when extracted from images acquired with TEs 5 ms apart (values decreased when increasing TE intervals) and 90.7% of the features exhibited excellent reproducibility for changes in TR. 2.0% of non-shape features were identified as providing only shape information. This study demonstrates that radiomic features are affected by specific MRI protocols. The use of our radiomic pelvic phantom allowed to identify unreliable features for radiomic analysis on T$_2$-weighted images. This paper proposes a general workflow to identify repeatable, reproducible, and informative radiomic features, fundamental to ensure robustness of clinical studies.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge