Alessandro Lascialfari
A kinetic approach to consensus-based segmentation of biomedical images
Nov 08, 2022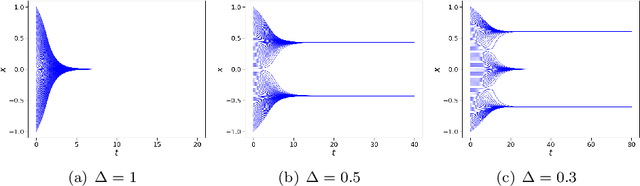
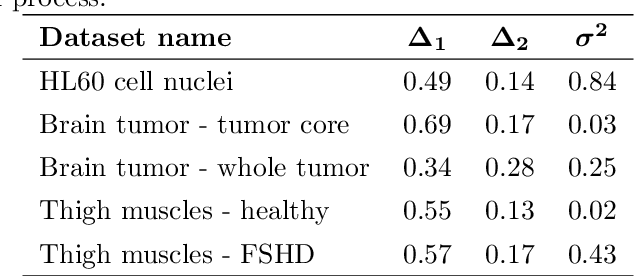
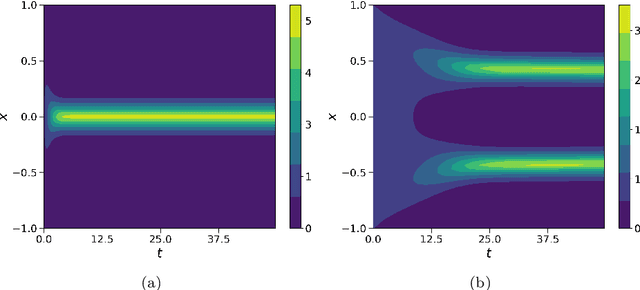
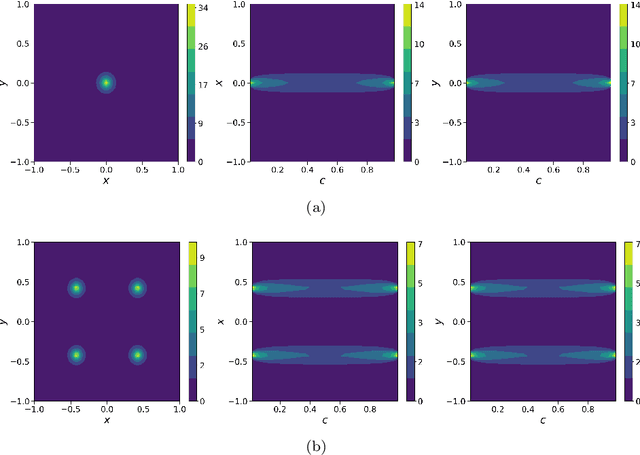
Abstract:In this work, we apply a kinetic version of a bounded confidence consensus model to biomedical segmentation problems. In the presented approach, time-dependent information on the microscopic state of each particle/pixel includes its space position and a feature representing a static characteristic of the system, i.e. the gray level of each pixel. From the introduced microscopic model we derive a kinetic formulation of the model. The large time behavior of the system is then computed with the aid of a surrogate Fokker-Planck approach that can be obtained in the quasi-invariant scaling. We exploit the computational efficiency of direct simulation Monte Carlo methods for the obtained Boltzmann-type description of the problem for parameter identification tasks. Based on a suitable loss function measuring the distance between the ground truth segmentation mask and the evaluated mask, we minimize the introduced segmentation metric for a relevant set of 2D gray-scale images. Applications to biomedical segmentation concentrate on different imaging research contexts.
Quantification of pulmonary involvement in COVID-19 pneumonia by means of a cascade oftwo U-nets: training and assessment on multipledatasets using different annotation criteria
May 06, 2021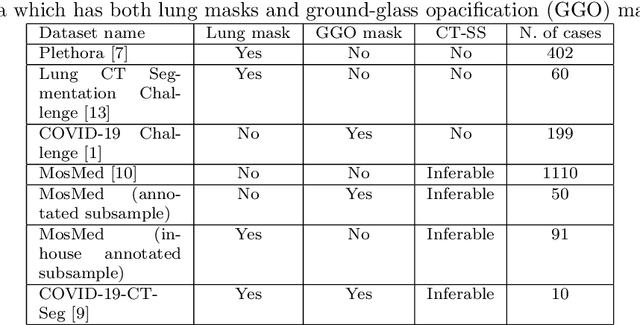
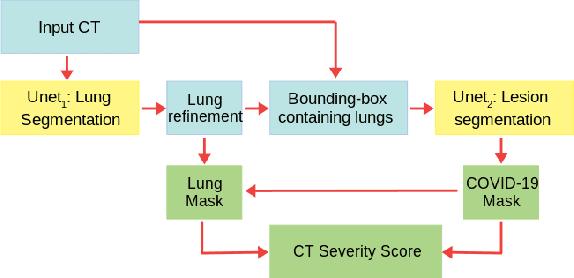
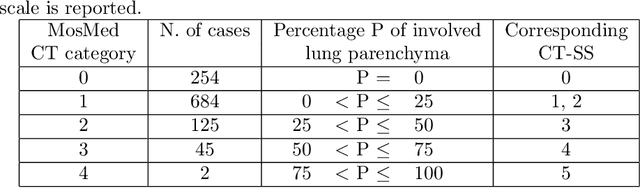
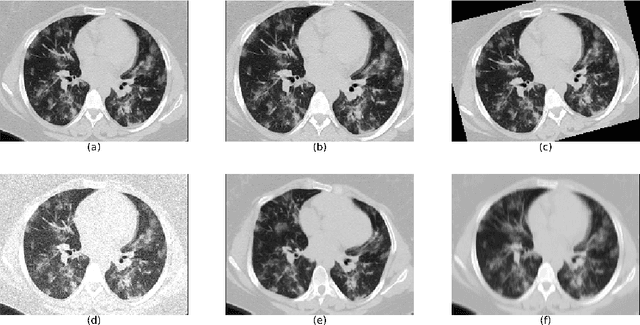
Abstract:The automatic assignment of a severity score to the CT scans of patients affected by COVID-19 pneumonia could reduce the workload in radiology departments. This study aims at exploiting Artificial intelligence (AI) for the identification, segmentation and quantification of COVID-19 pulmonary lesions. We investigated the effects of using multiple datasets, heterogeneously populated and annotated according to different criteria. We developed an automated analysis pipeline, the LungQuant system, based on a cascade of two U-nets. The first one (U-net_1) is devoted to the identification of the lung parenchyma, the second one (U-net_2) acts on a bounding box enclosing the segmented lungs to identify the areas affected by COVID-19 lesions. Different public datasets were used to train the U-nets and to evaluate their segmentation performances, which have been quantified in terms of the Dice index. The accuracy in predicting the CT-Severity Score (CT-SS) of the LungQuant system has been also evaluated. Both Dice and accuracy showed a dependency on the quality of annotations of the available data samples. On an independent and publicly available benchmark dataset, the Dice values measured between the masks predicted by LungQuant system and the reference ones were 0.95$\pm$0.01 and 0.66$\pm$0.13 for the segmentation of lungs and COVID-19 lesions, respectively. The accuracy of 90% in the identification of the CT-SS on this benchmark dataset was achieved. We analysed the impact of using data samples with different annotation criteria in training an AI-based quantification system for pulmonary involvement in COVID-19 pneumonia. In terms of the Dice index, the U-net segmentation quality strongly depends on the quality of the lesion annotations. Nevertheless, the CT-SS can be accurately predicted on independent validation sets, demonstrating the satisfactory generalization ability of the LungQuant.
A multicenter study on radiomic features from T$_2$-weighted images of a customized MR pelvic phantom setting the basis for robust radiomic models in clinics
May 18, 2020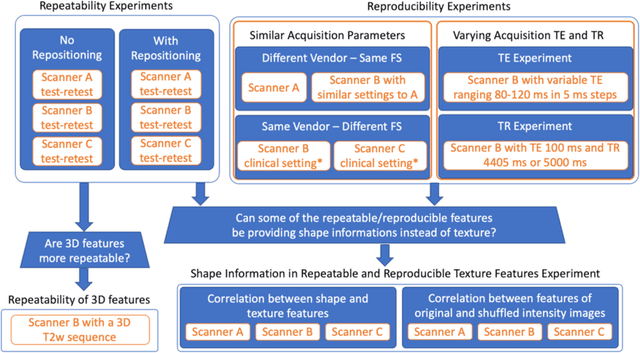
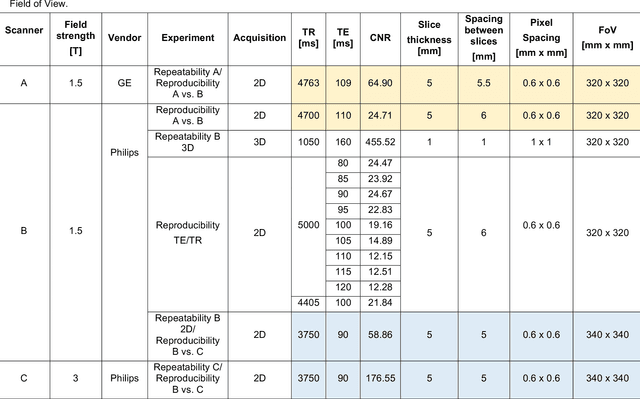
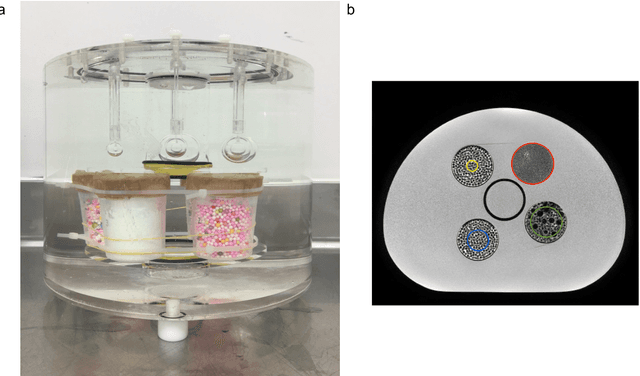
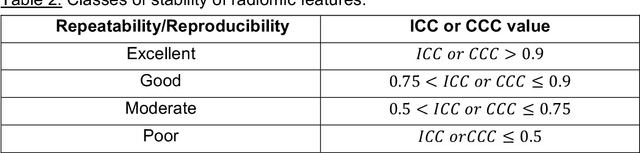
Abstract:In this study we investigated the repeatability and reproducibility of radiomic features extracted from MRI images and provide a workflow to identify robust features. 2D and 3D T$_2$-weighted images of a pelvic phantom were acquired on three scanners of two manufacturers and two magnetic field strengths. The repeatability and reproducibility of the radiomic features were assessed respectively by intraclass correlation coefficient (ICC) and concordance correlation coefficient (CCC), considering repeated acquisitions with or without phantom repositioning, and with different scanner/acquisition type, and acquisition parameters. The features showing ICC/CCC > 0.9 were selected, and their dependence on shape information (Spearman's $\rho$> 0.8) was analyzed. They were classified for their ability to distinguish textures, after shuffling voxel intensities. From 944 2D features, 79.9% to 96.4% showed excellent repeatability in fixed position across all scanners. Much lower range (11.2% to 85.4%) was obtained after phantom repositioning. 3D extraction did not improve repeatability performance. Excellent reproducibility between scanners was observed in 4.6% to 15.6% of the features, at fixed imaging parameters. 82.4% to 94.9% of features showed excellent agreement when extracted from images acquired with TEs 5 ms apart (values decreased when increasing TE intervals) and 90.7% of the features exhibited excellent reproducibility for changes in TR. 2.0% of non-shape features were identified as providing only shape information. This study demonstrates that radiomic features are affected by specific MRI protocols. The use of our radiomic pelvic phantom allowed to identify unreliable features for radiomic analysis on T$_2$-weighted images. This paper proposes a general workflow to identify repeatable, reproducible, and informative radiomic features, fundamental to ensure robustness of clinical studies.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge