Mario Masellis
Identifying latent disease factors differently expressed in patient subgroups using group factor analysis
Oct 10, 2024



Abstract:In this study, we propose a novel approach to uncover subgroup-specific and subgroup-common latent factors addressing the challenges posed by the heterogeneity of neurological and mental disorders, which hinder disease understanding, treatment development, and outcome prediction. The proposed approach, sparse Group Factor Analysis (GFA) with regularised horseshoe priors, was implemented with probabilistic programming and can uncover associations (or latent factors) among multiple data modalities differentially expressed in sample subgroups. Synthetic data experiments showed the robustness of our sparse GFA by correctly inferring latent factors and model parameters. When applied to the Genetic Frontotemporal Dementia Initiative (GENFI) dataset, which comprises patients with frontotemporal dementia (FTD) with genetically defined subgroups, the sparse GFA identified latent disease factors differentially expressed across the subgroups, distinguishing between "subgroup-specific" latent factors within homogeneous groups and "subgroup common" latent factors shared across subgroups. The latent disease factors captured associations between brain structure and non-imaging variables (i.e., questionnaires assessing behaviour and disease severity) across the different genetic subgroups, offering insights into disease profiles. Importantly, two latent factors were more pronounced in the two more homogeneous FTD patient subgroups (progranulin (GRN) and microtubule-associated protein tau (MAPT) mutation), showcasing the method's ability to reveal subgroup-specific characteristics. These findings underscore the potential of sparse GFA for integrating multiple data modalities and identifying interpretable latent disease factors that can improve the characterization and stratification of patients with neurological and mental health disorders.
ROOD-MRI: Benchmarking the robustness of deep learning segmentation models to out-of-distribution and corrupted data in MRI
Mar 11, 2022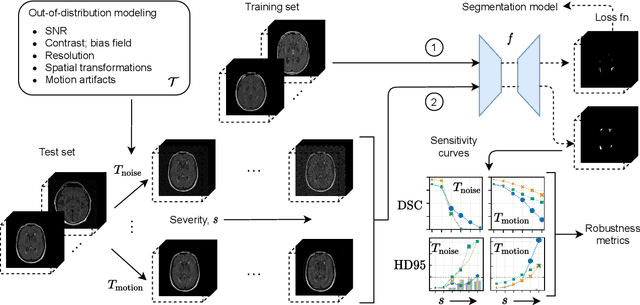
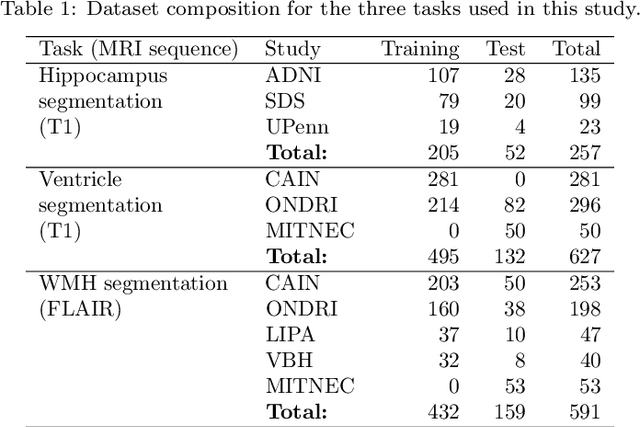
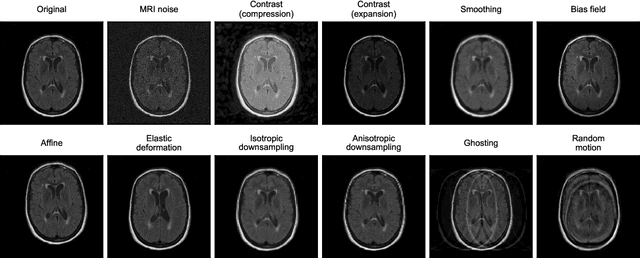
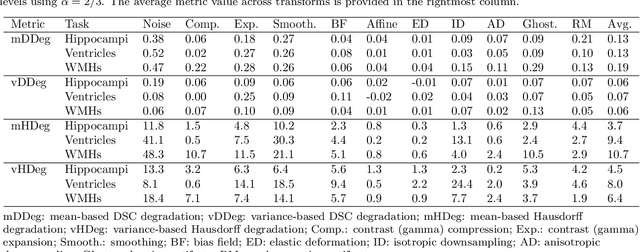
Abstract:Deep artificial neural networks (DNNs) have moved to the forefront of medical image analysis due to their success in classification, segmentation, and detection challenges. A principal challenge in large-scale deployment of DNNs in neuroimage analysis is the potential for shifts in signal-to-noise ratio, contrast, resolution, and presence of artifacts from site to site due to variances in scanners and acquisition protocols. DNNs are famously susceptible to these distribution shifts in computer vision. Currently, there are no benchmarking platforms or frameworks to assess the robustness of new and existing models to specific distribution shifts in MRI, and accessible multi-site benchmarking datasets are still scarce or task-specific. To address these limitations, we propose ROOD-MRI: a platform for benchmarking the Robustness of DNNs to Out-Of-Distribution (OOD) data, corruptions, and artifacts in MRI. The platform provides modules for generating benchmarking datasets using transforms that model distribution shifts in MRI, implementations of newly derived benchmarking metrics for image segmentation, and examples for using the methodology with new models and tasks. We apply our methodology to hippocampus, ventricle, and white matter hyperintensity segmentation in several large studies, providing the hippocampus dataset as a publicly available benchmark. By evaluating modern DNNs on these datasets, we demonstrate that they are highly susceptible to distribution shifts and corruptions in MRI. We show that while data augmentation strategies can substantially improve robustness to OOD data for anatomical segmentation tasks, modern DNNs using augmentation still lack robustness in more challenging lesion-based segmentation tasks. We finally benchmark U-Nets and transformer-based models, finding consistent differences in robustness to particular classes of transforms across architectures.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge